Transcription Factor-Mediated Gene Expression Regulation: Molecular Mechanisms and Biological Impact
A Comprehensive Analysis with Structural and Functional Insights
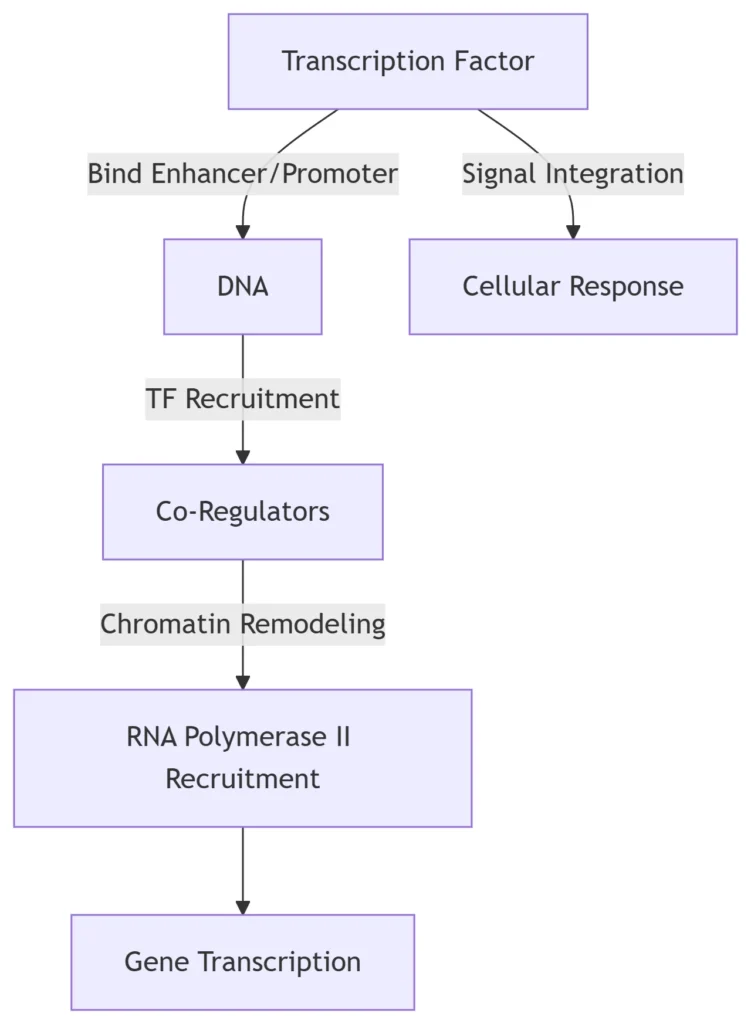
1. Introduction: Transcription Factors (TFs) as Genetic Switches
Transcription factors are DNA-binding proteins that recognize specific sequences (cis-elements) to activate or repress gene expression. They constitute ~8% of human genes and respond to developmental cues, environmental signals, and disease states through:
-
Sequence-specific DNA binding (nanomolar affinity)
-
Protein-protein interactions with co-regulators
-
Signal-responsive conformational changes
2. Core Regulatory Mechanisms
A. DNA Recognition Strategies
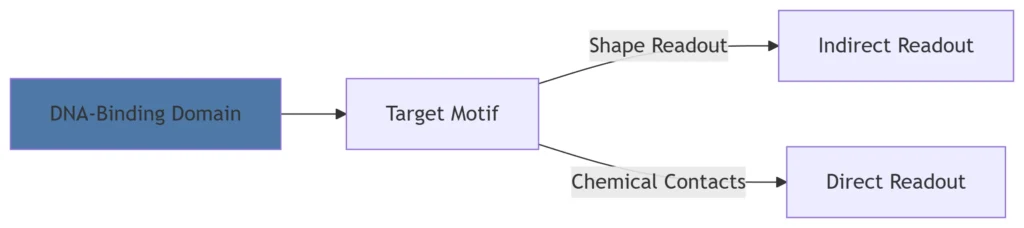
Key DNA-Binding Domains:
| Domain Type | Structure | Target Sequence | Example TF |
|---|---|---|---|
| Zinc Finger | ββα fold with Zn²⁺ | GC-rich boxes | SP1 |
| Helix-Turn-Helix | Two α-helices | Palindromic sites | p53 |
| Leucine Zipper | Parallel coiled coils | AP-1 site (TGACTCA) | c-Fos/c-Jun |
| Helix-Loop-Helix | Two helices + flexible loop | E-box (CANNTG) | MyoD |
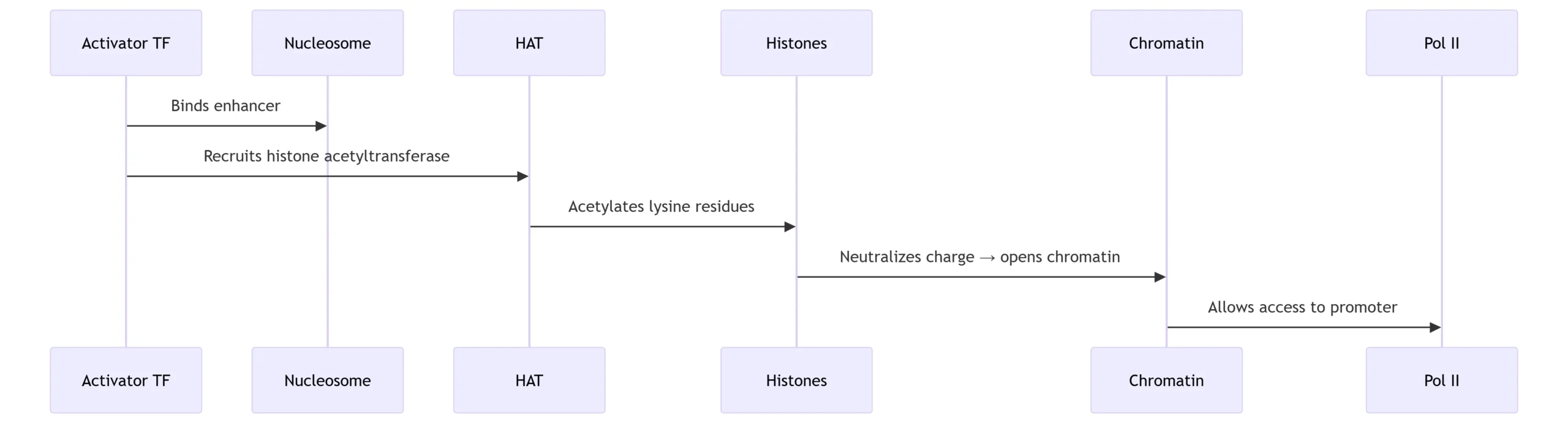
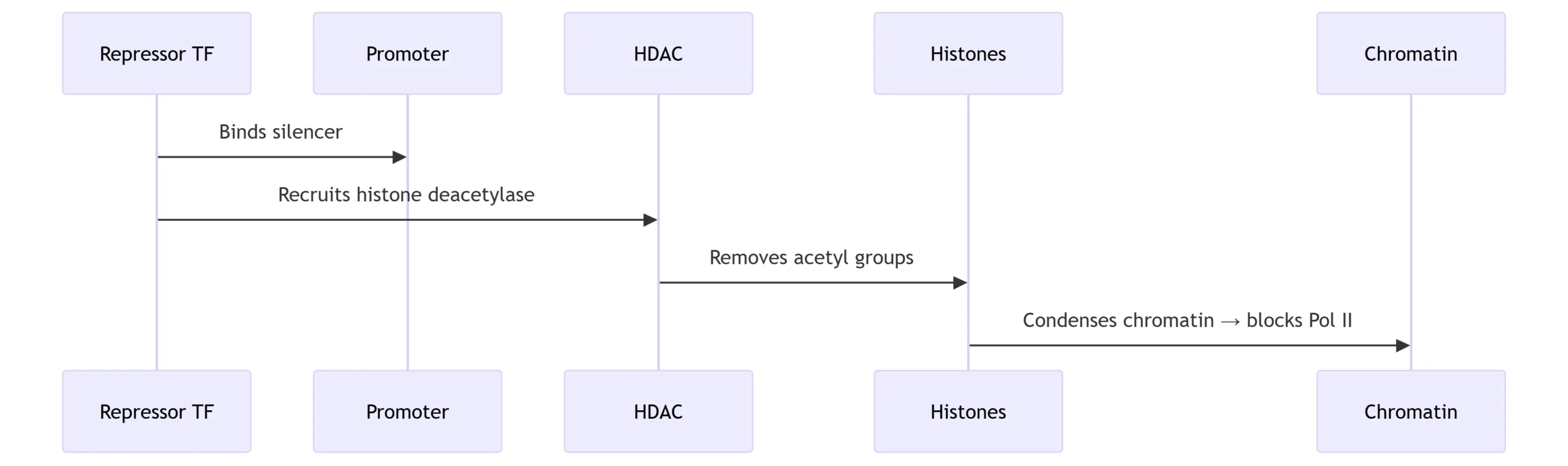
B. Chromatin Remodeling Pathways
Activation Mechanism:
Repression Mechanism:
3. TF Functional Domains
Modular Architecture:
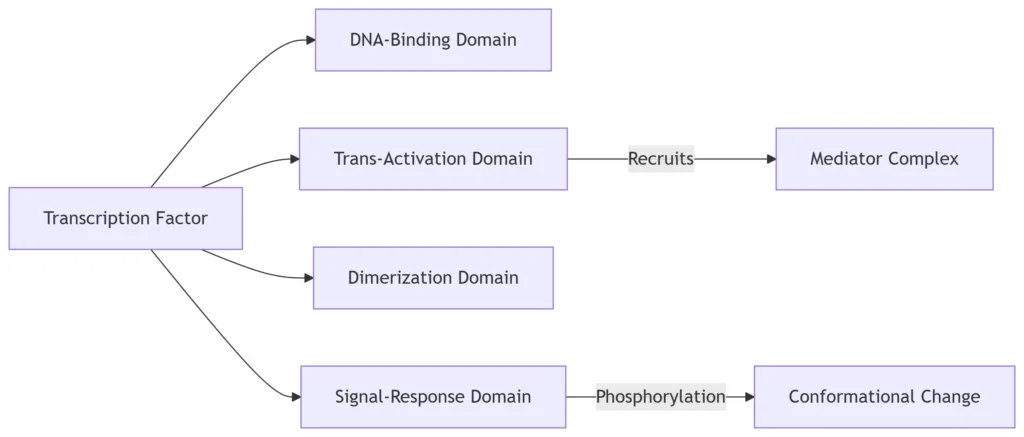
-
TAD interactions: Recruit RNA Pol II via Mediator (30-subunit complex)
-
SRD modifications: Phosphorylation, acetylation, ubiquitination alter TF activity
4. Combinatorial Control Principles
Enhancer Logic
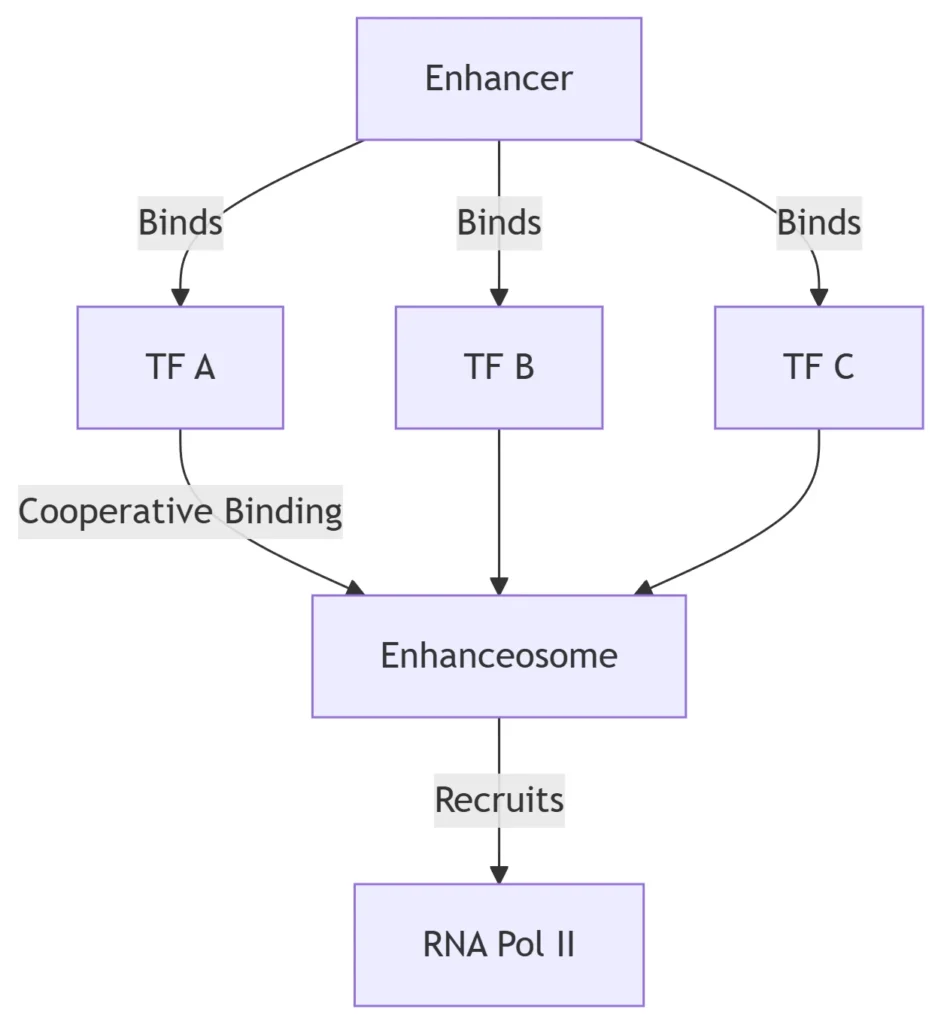
Key Features:
-
TF Cooperativity: Synergistic binding (e.g., NFAT:AP-1 at immune genes)
-
Enhancer-Promoter Looping: Mediated by cohesin/CTCF (loop range: 10kb-1Mb)
-
Phase Separation: TFs form liquid condensates to concentrate transcriptional machinery
5. Signal-Responsive Regulation
TF Activation Pathways
| Signal | TF | Activation Mechanism | Target Genes |
|---|---|---|---|
| Growth Factors | c-Myc | ERK phosphorylation | Cell cycle promoters |
| Stress | HSF1 | Trimerization & nuclear import | Heat shock proteins |
| Inflammation | NF-κB | IκB degradation | Cytokines/CAMs |
| Hormones | Estrogen R | Ligand-induced dimerization | Metabolic regulators |
NF-κB Example:

6. Quantitative Control Dynamics
TF Binding Kinetics:
| Parameter | Value | Functional Impact |
|---|---|---|
| Residence Time | 1-30 seconds | Determines burst frequency |
| On-Rate (k<sub>on</sub>) | 10⁴-10⁶ M⁻¹s⁻¹ | Affects signal sensitivity |
| Off-Rate (k<sub>off</sub>) | 0.1-10 s⁻¹ | Controls transcriptional noise |
Gene Expression Output:
-
Digital Control: All-or-nothing expression in single cells
-
Analog Control: Graded response to TF concentration
7. Regulatory Networks & Cellular Identity
Stem Cell Pluripotency Circuit
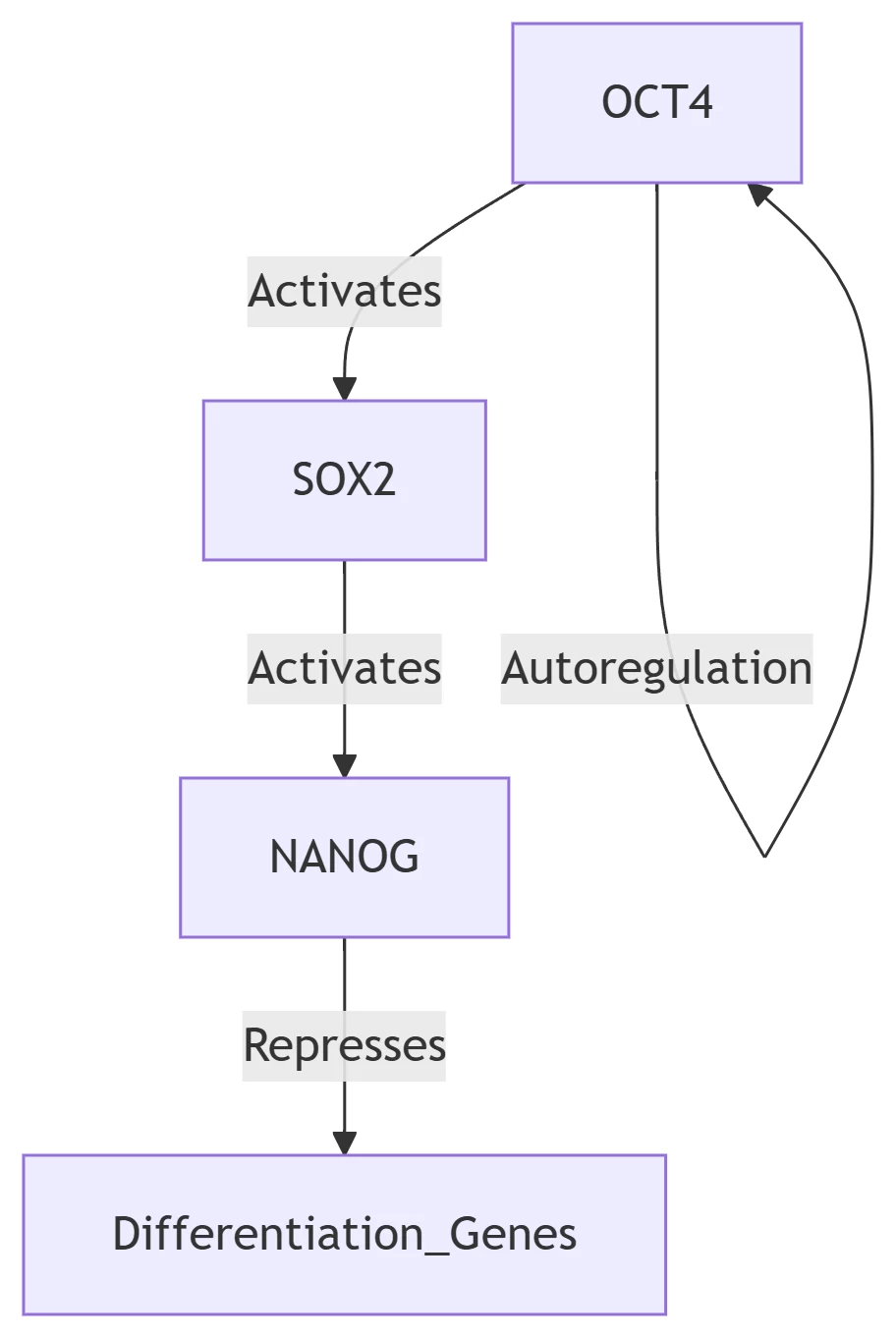
Network Properties:
-
Positive Feedback: Stabilizes pluripotent state
-
Mutual Exclusivity: TF cross-repression enables lineage commitment
8. Disease Implications & Therapeutics
Dysregulation Examples:
| Disease | TF Defect | Consequence |
|---|---|---|
| Breast Cancer | ERα overexpression | Uncontrolled proliferation |
| Diabetes | PDX1 deficiency | Impaired insulin production |
| Autoimmunity | FOXP3 mutation | T-reg dysfunction |
Therapeutic Strategies:
-
Small Molecules: Tamoxifen (blocks ERα-DNA binding)
-
PROTACs: Degrade oncogenic TFs (e.g., ARV-471 for ERα)
-
Gene Therapy: Restore TF function (e.g., FOXP3 in IPEX syndrome)
Conclusion
Transcription factors orchestrate gene expression through four-tiered regulation:
-
DNA Recognition: Sequence-specific binding via structural motifs
-
Chromatin Modulation: Recruitment of remodeling complexes (HAT/HDAC)
-
Machine Recruitment: Assembly of transcriptional machinery at promoters
-
Signal Integration: Post-translational modifications fine-tune activity
Their combinatorial control enables precise responses with <5% expression noise, while network architectures establish cellular identities. Dysregulation causes ~20% of human diseases, making TFs prime therapeutic targets. Emerging technologies (Chip-seq, single-cell ATAC) continue to decode TF regulatory grammars, advancing precision medicine.
Data sourced from public references including:
-
Lambert S.A. et al. The Human Transcription Factors (Cell, 2018)
-
Vaquerizas J.M. Nature Reviews Genetics (2009)
-
ENCODE TF ChIP-seq Database
-
RCSB PDB structures: 1TUP (p53), 2HZD (NF-κB)
For academic collaboration or content inquiries: chuanchuan810@gmail.com
