A Comprehensive Exploration of Transcription Dynamics Across Life Domains
1. Introduction
RNA polymerase (RNAP) is the central enzymatic machinery driving transcription, the process by which genetic information encoded in DNA is transcribed into RNA. This foundational step in gene expression enables the synthesis of functional RNA molecules—messenger RNA (mRNA), ribosomal RNA (rRNA), transfer RNA (tRNA), and non-coding RNAs—that orchestrate cellular activities. RNAP operates universally across prokaryotes, eukaryotes, and archaea, with structural and functional adaptations tailored to biological complexity. This article elucidates RNAP’s role in transcription, its evolutionary conservation, and its implications for biotechnology and medicine.
2. Structural Architecture of RNA Polymerase
A. Prokaryotic RNA Polymerase
Prokaryotic RNAP is a multi-subunit complex (α₂ββ’ω) coupled with a dissociable sigma (σ) factor that confers promoter specificity. The σ factor directs RNAP to conserved promoter elements, such as the -35 hexamer and -10 Pribnow box, ensuring precise initiation . The core enzyme (α₂ββ’ω) facilitates RNA synthesis, with the β and β’ subunits forming a clamp-like structure that stabilizes DNA interactions during elongation .
B. Eukaryotic RNA Polymerase
Eukaryotes possess three distinct RNA polymerases:
- RNAP I: Transcribes rRNA precursors (e.g., 45S pre-rRNA) in the nucleolus.
- RNAP II: Synthesizes mRNA and regulatory RNAs (e.g., miRNA, snRNA), requiring a TATA box and transcription factors (e.g., TFIID) for promoter recognition. RNAP II’s C-terminal domain (CTD) coordinates RNA processing through dynamic phosphorylation .
- RNAP III: Produces tRNA, 5S rRNA, and other small RNAs.
Suggested Figure: Comparative 3D models of prokaryotic (σ-bound) and eukaryotic RNAP II, highlighting subunit composition and functional domains.
3. The Transcription Cycle: Initiation, Elongation, and Termination
A. Initiation: Promoter Recognition and Open Complex Formation
Transcription begins with RNAP binding to promoter regions. In prokaryotes, the σ factor guides RNAP to the promoter, while in eukaryotes, transcription factors (e.g., TFIID for RNAP II) mediate this process . DNA unwinding forms a transcription bubble, exposing the template strand for RNA synthesis.
Suggested Figure: Initiation complex showing RNAP bound to promoter DNA, with σ factor (prokaryotes) or TFIID (eukaryotes).
B. Elongation: RNA Chain Polymerization
RNAP catalyzes RNA synthesis via phosphodiester bond formation between ribonucleoside triphosphates (NTPs), guided by complementary base pairing (A-U, G-C). The enzyme moves processively along DNA, maintaining a transcription bubble and rewinding DNA upstream . Unlike DNA polymerases, RNAP lacks proofreading activity, resulting in an error rate of ~10⁻⁴ .
Suggested Figure: Catalytic site of RNAP during elongation, illustrating NTP incorporation and transcription bubble dynamics.
C. Termination: Release of RNA and Enzyme Dissociation
- Prokaryotes: Termination occurs via intrinsic (hairpin structures and poly-U tracts) or ρ-dependent (helicase-mediated) mechanisms .
- Eukaryotes: RNAP II termination involves polyadenylation signals and cofactors (e.g., CPSF), which cleave the transcript and recruit exonucleases .
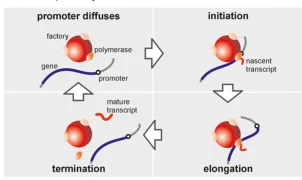
Suggested Figure: Termination mechanisms: hairpin-induced dissociation (prokaryotes) vs. polyadenylation signal recognition (eukaryotes).
4. Transcriptional Regulation and Functional Plasticity
A. Prokaryotic Regulation
- Sigma Factor Switching: E. coli employs alternative σ factors (e.g., σ³² under heat shock) to redirect RNAP to stress-responsive promoters .
- Transcription Attenuation: RNAP pausing regulates operons (e.g., trp operon) by coupling transcription to metabolite availability.
B. Eukaryotic Complexity
- Enhancers and Silencers: Distal DNA elements loop to promoters via mediator complexes, modulating RNAP II activity .
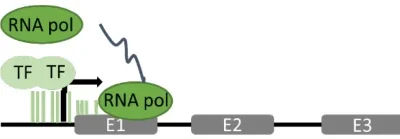
- Epigenetic Control: Histone modifications (e.g., acetylation) and DNA methylation regulate RNAP accessibility to genes .
Suggested Figure: Regulatory network of eukaryotic RNAP II, showing enhancer-promoter looping and chromatin remodeling.
5. Evolutionary Insights and Structural Conservation
RNAP’s core subunits (β, β’, α) are highly conserved, reflecting its essential role in gene expression. Archaeal RNAP shares features with both eukaryotes and bacteria, offering insights into evolutionary divergence . Structural studies of yeast RNAP II at 2.8 Å resolution reveal mobile modules (e.g., the clamp) that mediate DNA entry and RNA exit, underscoring mechanistic universality .
6. Applications in Biotechnology and Medicine
A. Antibiotic Targets
Bacterial RNAP is targeted by antibiotics like rifampicin, which blocks the RNA exit channel to treat tuberculosis .
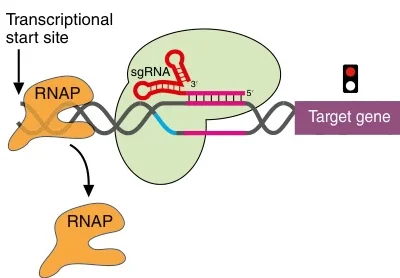
B. CRISPR-Cas9 Systems
CRISPR guide RNAs are transcribed by RNAP, enabling precise genome editing in gene therapy and synthetic biology .
C. Disease Mechanisms and Therapies
- Premature Termination Codons (PTCs): Readthrough drugs (e.g., ataluren) bypass PTCs in diseases like cystic fibrosis .
- Cancer and Neurodevelopmental Disorders: RNAP II mutations are linked to transcriptional dysregulation, driving therapeutic innovation .
Suggested Figure: CRISPR-Cas9 system utilizing RNAP-transcribed guide RNAs for gene editing.
7. Challenges and Future Directions
- Structural Dynamics: Cryo-EM and single-molecule studies are resolving RNAP’s conformational changes during transcription .
- AI-Driven Design: Machine learning models predict promoter-RNAP interactions, accelerating synthetic biology applications .
- Therapeutic Targeting: Developing RNAP inhibitors for viral infections and cancer remains a frontier .
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com
