 I. Genomic Identity and Translation Initiation
I. Genomic Identity and Translation Initiation
Positive-sense RNA (+ssRNA) genomes function as immediate messenger RNA (mRNA) upon host cell entry, with their nucleotide sequence directly recognized by host ribosomes for instantaneous protein synthesis. This eliminates the need for viral transcription machinery prior to translation initiation .
(Fig. 1: Direct Translation Mechanism)
Description: Ribosome (grey) binding to +ssRNA genome (blue) at the 5′ untranslated region (UTR), initiating synthesis of viral polyproteins. The 3′ UTR contains conserved stem-loop structures facilitating ribosome recruitment.
II. Molecular Workflow of Early Infection
Step 1: Primary Translation
- First-Responder Proteins: The +ssRNA genome directs synthesis of RNA-dependent RNA polymerase (RdRp) and replication cofactors within minutes of cellular entry .
- Host Machinery Hijacking: Uses host tRNAs, amino acids, and energy (ATP/GTP) for translation, bypassing nuclear processing .
Step 2: Replication Complex Assembly
- Membrane Remodeling: RdRp recruits host proteins to reshape endoplasmic reticulum (ER) membranes into vesicle clusters (double-membrane vesicles, DMVs). These protect dsRNA intermediates from host immune sensors .
- Template Recruitment: Viral cis-acting RNA elements (e.g., stem-loops in coronaviruses) direct genome localization to DMVs .
(Fig. 2: Replication Organelle Formation)
Description: ER membrane (yellow) invaginating to form DMVs (spheres). +ssRNA (blue) enters vesicles where RdRp (purple) synthesizes complementary strands.
III. Replication-Translation Switch
| Process | Molecular Mechanism | Regulatory Elements |
|---|---|---|
| Translation Phase | Cap-dependent ribosomal scanning | 5′ cap structure, IRES elements |
| Replication Phase | RdRp binding to 3′ UTR | Stem-loop pseudoknots, packaging signals |
| Transition Trigger | Accumulation of viral proteases | Cleavage of host eIF4G factor |
Critical Balance:
- Competition Avoidance: The same +ssRNA molecule cannot simultaneously serve as translation template and replication substrate. Host RNA-binding proteins (e.g., LSm1-7 complex) regulate access .
IV. Asymmetric Amplification Cycle
- (-) Strand Synthesis: RdRp uses +ssRNA as template to generate complementary (-)RNA .
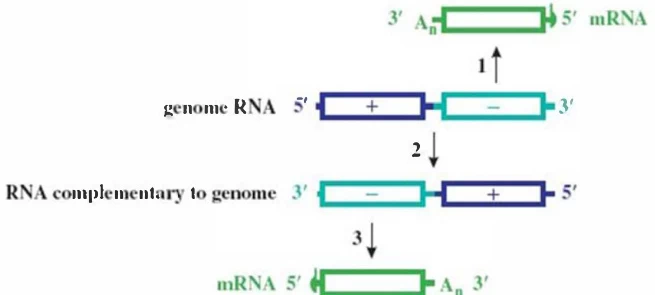
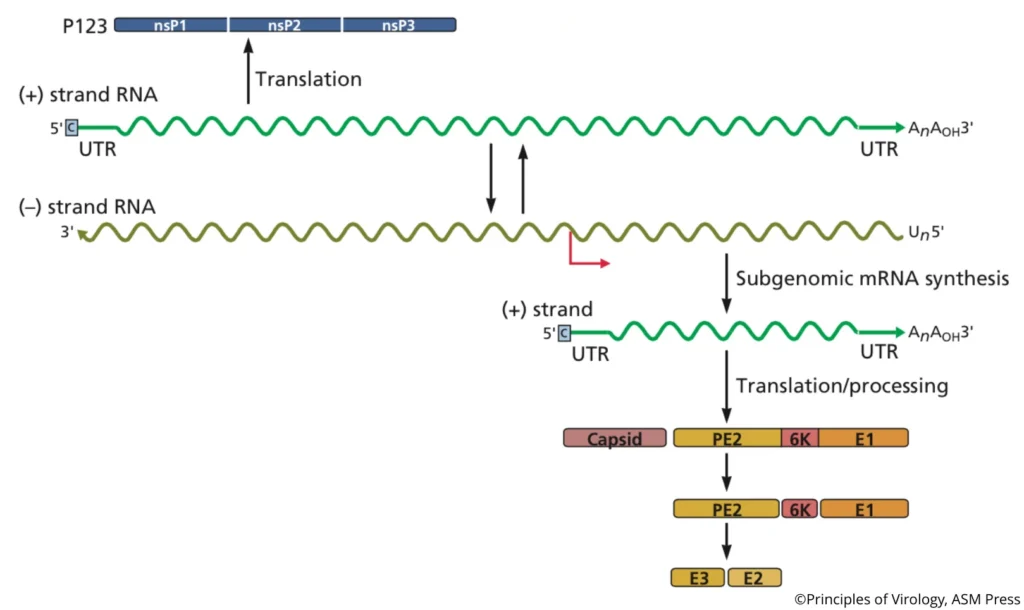
- Replication Intermediate: Forms transient dsRNA (replicative form, RF) within DMVs .
- Progeny Production: (-)RNA template generates 10-100x more (+)ssRNA than genomic copies:
- 30% → New genomes
- 70% → Subgenomic mRNAs for structural proteins
(Fig. 3: Replication Asymmetry)
Description: RdRp (purple) bound to (-)RNA template (red) producing multiple +ssRNA strands (blue). Subgenomic mRNAs (shorter blue fragments) translate capsid proteins.
V. Clinical & Evolutionary Implications
A. Pathogenesis Advantages
- Rapid Onset: Translation within 10 minutes enables immune evasion (e.g., SARS-CoV-2 suppresses IFN within 4 hrs) .
- High Mutation Rates: Lack of proofreading creates quasispecies diversity, facilitating host adaptation .
B. Therapeutic Targets
| Target | Inhibitor Example | Mechanism |
|---|---|---|
| 5′ Capping | Ribavirin | Impairs translation initiation |
| RdRp Active Site | Remdesivir | Blocks (-) strand synthesis |
| DMV Formation | Cyclophilin inhibitors | Prevents replication organelle assembly |
VI. Synthetic Biology Applications
A. mRNA Vaccine Platforms
- Self-Amplifying RNA (saRNA): Engineered alphavirus replicons encoding antigens (e.g., COVID-19 vaccines), boosting expression 50x vs. conventional mRNA .
- Rational Design: Codon-optimized 5′ UTRs enhance ribosomal engagement; modified 3′ UTRs extend half-life .
B. Gene Therapy Vectors
- Replicon Systems: +ssRNA replicons delivering therapeutic genes (e.g., Factor IX for hemophilia) with sustained expression .
“+ssRNA viruses exemplify genomic economy: a single molecule serves as infectious agent, mRNA template, and replication blueprint—rewriting central dogma constraints.”
— Cell, 2024
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
