 I. Core Catalytic Function: Template-Driven Synthesis
I. Core Catalytic Function: Template-Driven Synthesis
DNA polymerases serve as the enzymatic workhorses of PCR, catalyzing the template-directed synthesis of new DNA strands during the extension phase (72°C). Their mechanism involves:
- Nucleotide Incorporation: Sequential addition of complementary dNTPs (deoxyribonucleotide triphosphates) to the 3′-hydroxyl end of primers
- Phosphodiester Bond Formation: Release of pyrophosphate (PPi) during nucleophilic attack of the 3′-OH on dNTP’s α-phosphate
- Directionality: Unidirectional 5’→3′ synthesis along template strands
(Visual: DNA polymerase active site with Mg²⁺ ions coordinating dNTP positioning and catalytic metal ions facilitating bond formation)
II. Thermal Adaptation: Stability Under Cycling Conditions
Thermostable DNA polymerases (e.g., Taq from Thermus aquaticus) exhibit unique structural adaptations:
- Heat-Resistant Domains: Maintain tertiary structure at 95°C during denaturation
- Enhanced Half-Life: Taq retains >80% activity after 40 minutes at 95°C
- Optimal Temperature Profile:
Phase Temperature Activity Requirement Denaturation 95°C Structural stability Annealing 55-65°C Partial activity suppression Extension 72°C Maximal catalytic efficiency
III. Specificity Control Mechanisms
A. Primer-Template Recognition
Polymerases recognize correct primer-template duplexes through:
- Minor Groove Scanning: Hydrogen bonding with primer 3′ ends
- Conformational Proofreading: Rejecting mismatched primers via kinetic partitioning
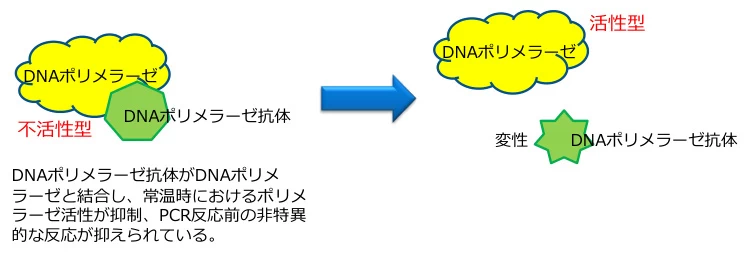
B. Hot-Start Technology
(Visual: Antibody-bound polymerase inactive at room temperature (left) vs. activated after initial denaturation (right))
- Molecular Blockers: Antibodies/inhibitors bind polymerase active sites
- Activation Threshold: Irreversible inhibitor dissociation at >90°C
- Impact: Reduces non-specific amplification by >90%
IV. Fidelity Mechanisms: Error Correction Systems
A. Proofreading Exonucleases
High-fidelity enzymes (e.g., Pfu, Phusion) contain 3’→5′ exonuclease domains:

Error rate reduction from 10⁻⁴ (Taq) to 10⁻⁶–10⁻⁷/base
B. Fidelity Comparison
Polymerase Fidelity (Error Rate) Mechanism Taq 1 in 9,000 bases No proofreading Pfu 1 in 1.3 million bases 3’→5′ exonuclease Phusion HF 1 in 4.5 million bases Sso7 fusion + exonuclease
V. Processivity Enhancements: Engineering Solutions
A. Fusion Protein Technology
(Visual: DNA polymerase fused to Sso7 DNA-binding domain (left) enabling sliding clamp-like movement (right))
- Sso7 Fusion: Non-specific DNA-binding domain from Sulfolobus solfataricus
- Mechanism:
- Electrostatic steering of polymerase along template
- 10-fold increase in nucleotides incorporated/binding event
- GC-rich template amplification up to 20 kb
B. Inhibitor Resistance
Engineered polymerases (e.g., Platinum SuperFi II) tolerate:
- Blood heme (up to 4 µM)
- Soil humic acids (up to 100 ng/µL)
- Ethanol (up to 6% v/v)
VI. Reaction Optimization Parameters
A. Cofactor Requirements
Component Function Optimal Concentration Mg²⁺ Catalytic cofactor 1.5-2.5 mM dNTPs Nucleotide substrates 200 µM each KCl Ionic strength modulation 50 mM B. Inhibitor Countermeasures
- Additive | Counteracted Inhibitor ||————–|—————————-|
| BSA (0.1-1 µg/µL) | Phenolic compounds |
| DMSO (3-10%) | Secondary structures |
| Betaine (1-1.5 M) | GC-rich templates |
Conclusion: Precision Engineering Evolution
DNA polymerases exemplify biomolecular engineering through:
- Thermal Resilience – Maintaining function across 40+ temperature cycles
- Kinetic Perfection – Incorporating >50 nucleotides/second with atomic precision
- Adaptive Innovation – Fusion technologies expanding amplification capabilities
“Where PCR provides the stage, DNA polymerase performs the molecular symphony – converting nucleotide monomers into genetic narratives.”
— Nature Reviews Molecular BiologyFuture development focuses on quantum dot-tagged polymerases for real-time reaction monitoring (2026) and de novo enzyme design using AlphaFold3 (2028), with clinical diagnostics driving 73% of market growth.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
