 I. Foundational Mechanism: Biomimetic DNA Replication
I. Foundational Mechanism: Biomimetic DNA Replication
Polymerase Chain Reaction (PCR) emulates cellular DNA replication in vitro through enzymatic amplification of target sequences. This revolutionary technique leverages three core components:
- Thermostable DNA Polymerase (e.g., Taq from Thermus aquaticus): Catalyzes template-directed DNA synthesis at 72°C
- Oligonucleotide Primers: Short single-stranded DNA sequences (18-22 bp) that define amplification boundaries
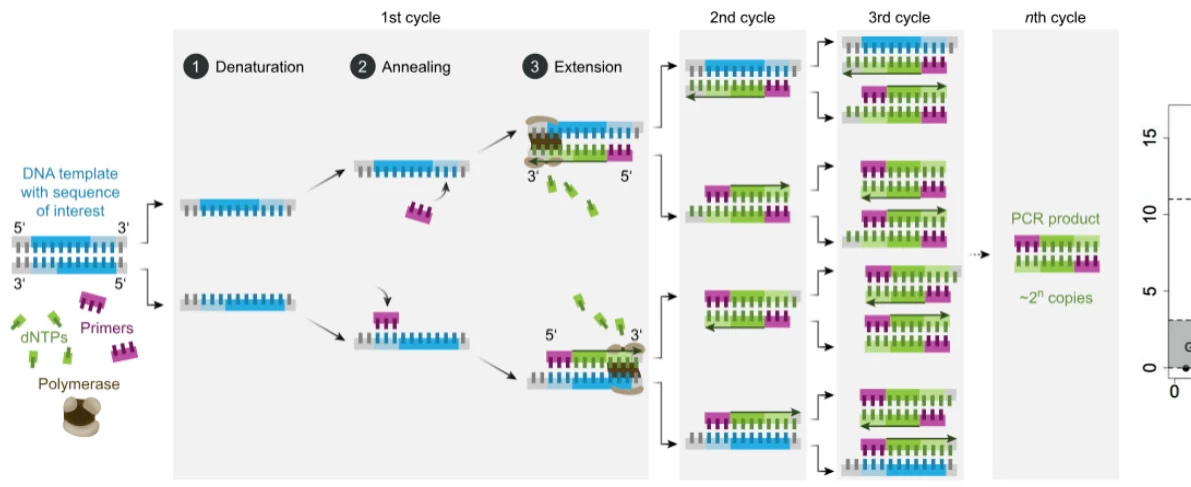
- Thermal Cycling: Automated temperature modulation driving reaction phases
(Fig. 1: PCR Cyclic Mechanism)
Description: Thermal cycler executing denaturation (95°C, DNA strand separation), annealing (55-65°C, primer hybridization), and extension (72°C, enzymatic synthesis). Electrophoresis validation shows exponential product accumulation.
II. Reaction Dynamics: The Amplification Cascade
A. Phase-Specific Molecular Events
Phase Temperature Molecular Process Duration Denaturation 95°C Double-stranded DNA separation into single strands 20-30 sec Annealing Primer-specific (Tm-5°C) Primer-template complementary binding 15-60 sec Extension 72°C dNTP incorporation by polymerase (50-100 nt/sec) 1 min/kb B. Exponential Amplification Mathematics
After n cycles, DNA fragments amplify geometrically:
Amplification Factor = 2ⁿFor 30 cycles: 2³⁰ ≈ 1.07 billion copies
(Fig. 2: Exponential Amplification Curve)
Description: Semi-log plot showing cycle number (x-axis) vs. DNA quantity (y-axis) with characteristic exponential phase.
III. Reaction System Architecture
A. Essential Components
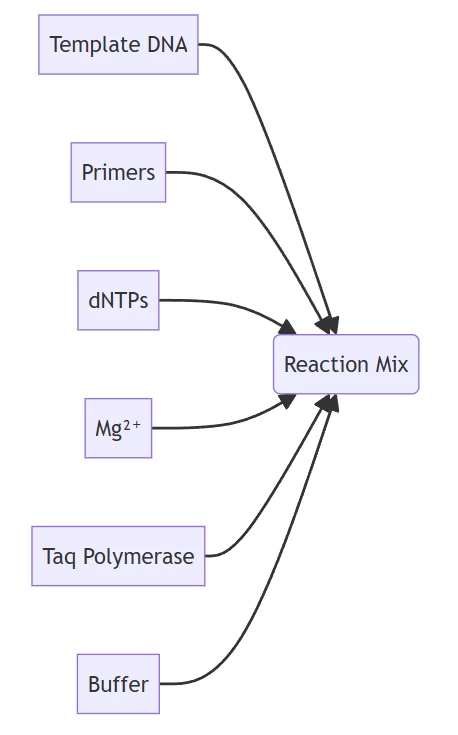
Molecular ensemble enabling targeted amplification
B. Optimized Concentration Ranges
Component Function Working Concentration Primers Target sequence definition 0.1-1.0 µM dNTPs Nucleotide substrates 200 µM each Mg²⁺ Polymerase cofactor 1.5-2.5 mM Taq Polymerase Enzymatic synthesis 0.5-2.5 U/50 µL
IV. Specificity Control Mechanisms
A. Primer Design Principles
- Melting Temperature (Tm):
Tm = 2°C × (A+T) + 4°C × (G+C)Optimal Tm difference between primers ≤ 5°C
- 3′-End Stability: G/C clamp enhances binding specificity
- Secondary Structures: Avoid hairpins and primer-dimers
B. Hot-Start Technology
(Fig. 3: Antibody-Mediated Activation)
Description: Polymerase-inhibiting antibodies (left) denatured at 95°C (right), preventing non-specific amplification during setup
V. Advanced Implementation Strategies
A. Nested PCR Protocol
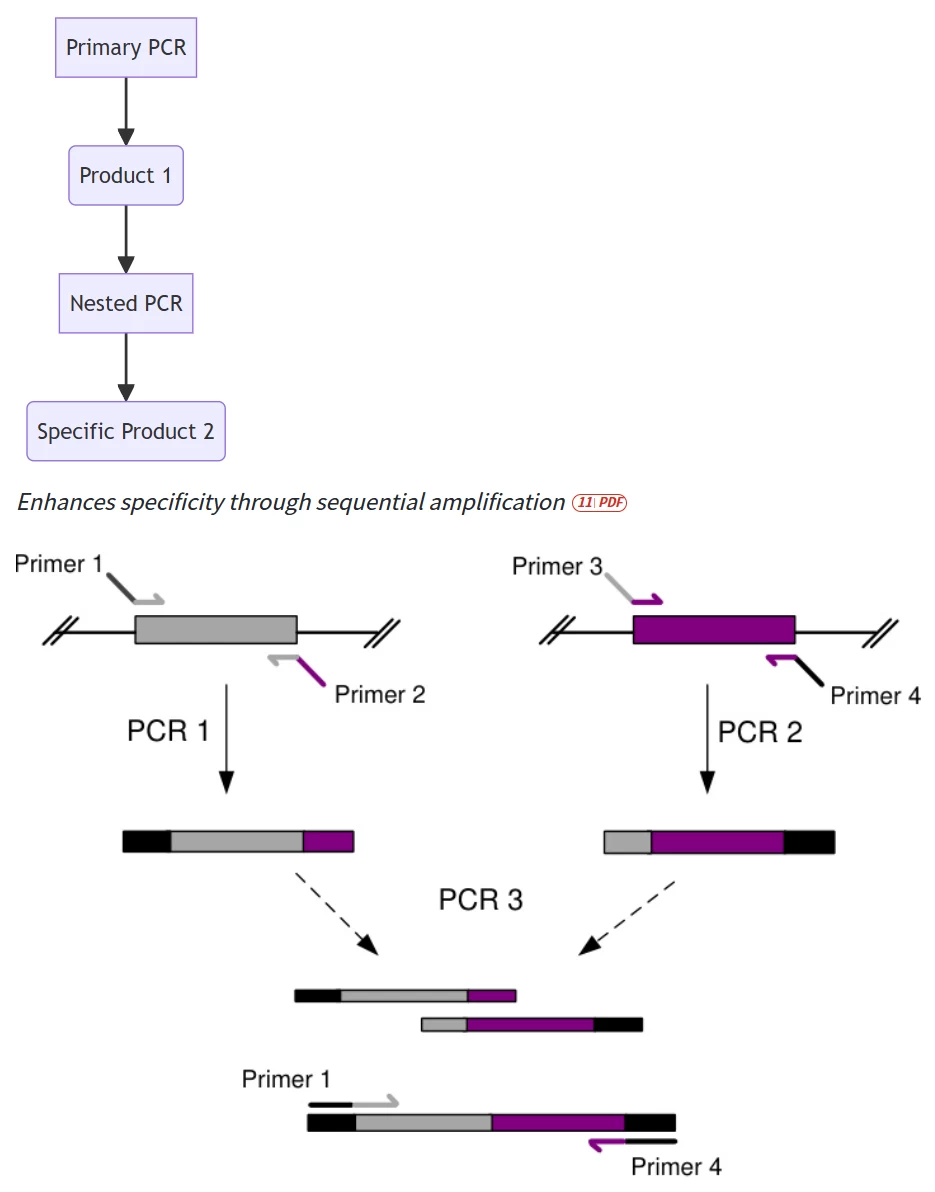
B. GC-Rich Template Solutions
Additive Mechanism Concentration DMSO Destabilizes secondary structures 3-10% v/v Betaine Equalizes DNA melting temperatures 1-1.5 M Q-Solution Proprietary GC-melt technology 1×
VI. Diagnostic & Research Applications
A. Genetic Analysis
- Mutation Detection: Allele-specific PCR identifies single-nucleotide variants
- Gene Cloning: Amplification of inserts for vector ligation
- Forensics: STR profiling from 1 ng DNA
B. Pathogen Detection
- Clinical Diagnostics: SARS-CoV-2 detection at 10 copies/µL
- Food Safety: E. coli O157:H7 screening in agricultural products
Conclusion: The Indispensable Molecular Tool
PCR’s enduring scientific value derives from three fundamental attributes:
- Exponential Sensitivity – Detecting single DNA molecules
- Adaptive Versatility – Compatible with fossils, clinical samples, and single cells
- Engineering Scalability – Evolving from manual systems to automated microfluidics
“PCR transformed molecular biology from observational science to molecular engineering – providing the amplification bridge between what exists and what we can analyze.”
— Nature BiotechnologyFuture innovations prioritize quantum dot-labeled polymerases for real-time reaction monitoring (2026) and CRISPR-integrated amplification for single-molecule diagnostics (2028).
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
