 I. Defining the Evolimb Research Field
I. Defining the Evolimb Research Field
Evolutionary Developmental Biology (Evo-Devo) constitutes the core discipline investigating limb evolution, integrating:
- Deep Homology Principles:
- Conservation of Hox gene networks (Hoxd13, Hoxa13) across fish fins and tetrapod limbs
- Shared Shh signaling pathways in limb polarization across vertebrates
- Developmental Plasticity:
- Modular cis-regulatory evolution enabling limb diversification without protein-coding changes
- Tissue self-organization mechanisms driving skeletal patterning
(Fig. 1: Hoxd13 expression in zebrafish fin vs. mouse limb bud)
Description: Fluorescent in situ hybridization showing conserved expression domains (purple) defining proximal-distal axes.
II. Core Research Pillars
A. Evolutionary Genomics
| Research Focus | Key Mechanisms | Model Systems |
|---|---|---|
| Regulatory Landscape | Enhancer co-option, structural variants | Iberian mole (Talpa occidentalis) |
| Extreme Adaptation | Digit elongation, webbing retention | Bats (Chiroptera) |
| Limb Loss | Tbx suppression, Gli3 disruption | Snakes, limb-reduced lizards |
B. Developmental Dynamics
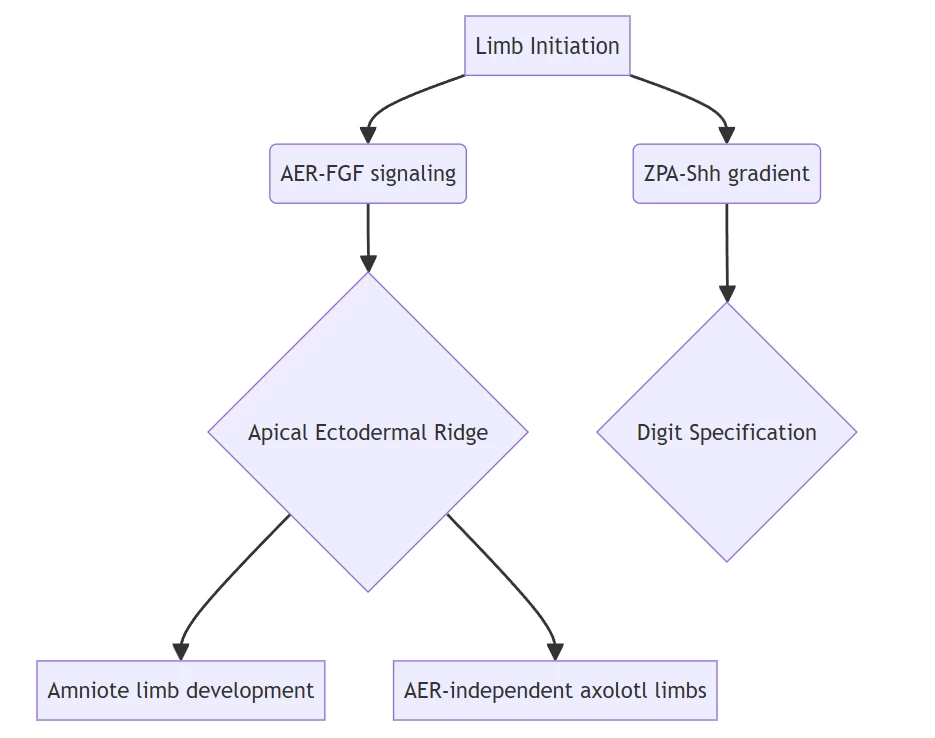
Evidence: Axolotls bypass AER requirements through mesenchymal Fgf expression
C. Paleontological Validation
- Transitional fossils (Eusthenopteron → Acanthostega) revealing fin-to-limb homology
- Whale pelvic reduction mirroring embryonic Hand2 suppression
(Fig. 2: Fossil sequence of cetacean limb reduction)
Description: Micro-CT reconstructions showing progressive pelvic girdle loss in Pakicetus → Ambulocetus → Dorudon.
III. Cutting-Edge Methodologies
1. Cross-Species Engineering
| Technique | Application | Discovery |
|---|---|---|
| CRISPR-Cas9 | Bat Prx1 enhancer in mice | Induced forelimb elongation |
| Single-cell ATAC-seq | Human limb cell atlas | Spatiotemporal gene regulatory networks |
| Paleoproteomics | Acanthostega collagen | Deep evolutionary conservation |
2. Mathematical Modeling
- Turing reaction-diffusion systems simulating digit patterning
- Quantitative trait evolution frameworks predicting limb morphology
IV. Key Unifying Concepts
The Evo-Devo Triad
- Deep Homology
- Ancient Hox–Shh axis predating tetrapods
- Developmental Bias
- Physical constraints on bone/joint formation
- Regulatory Rewiring
- Bat Fgf8 enhancer amplification enabling wing elongation
“Limbs are evolutionary palimpsests—each adaptation overwrites developmental instructions while preserving genomic foundations.”
– Synthesis of Evo-Devo Principles
(Fig. 3: Single-cell transcriptome map of human embryonic limb)
Description: Spatial transcriptomics identifying chondrogenic (blue), myogenic (red), and vascular (green) lineages.
V. Emerging Frontiers
1. Regenerative Medicine
- Axolotl limb regeneration mechanisms:
- Bmp4–Fgf20 axis reactivation
- Bioelectric prepattern memory
2. Human Clinical Relevance
- SYNPO2 enhancer deletions → syndactyly
- Gli3 regulatory mutations → polydactyly
3. Cross-Phylum Synthesis
- Arthropod vs. vertebrate limb evolution:
- Convergent joint formation via Bmp signaling
- Homologous segmentation logic
Conclusion: The Evo-Devo Synthesis
Evolimb research resides at the convergence of three disciplines:
- Evolutionary Biology: Fossil transitions + comparative genomics
- Developmental Genetics: Cell fate specification + signaling pathways
- Systems Biology: Multi-scale modeling + omics integration
“To study limb evolution is to decode nature’s most elegant morphogenetic algorithm—where deep conservation enables radical innovation.”
Future research will unravel how cis-regulatory grammar generates morphological diversity while advancing regenerative therapies.
Data sourced from publicly available references. For collaboration or domain interest inquiries, contact: chuanchuan810@gmail.com.
