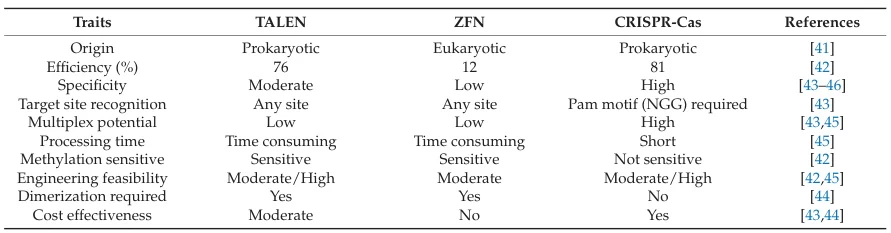
 I. Molecular Recognition Mechanisms: The Specificity Foundation
I. Molecular Recognition Mechanisms: The Specificity Foundation
A. ZFN: Zinc Finger Protein-DNA Binding
Zinc Finger Nucleases (ZFNs) combine zinc finger proteins (ZFPs) with FokI endonucleases. Each zinc finger module recognizes 3-4 base pairs via α-helix-DNA major groove interactions. ZFNs require dimerization to cleave DNA between 5-6 bp spacer regions. Their specificity is constrained by cross-talk between zinc fingers, where adjacent modules influence binding accuracy, leading to moderate off-target risks.
(Fig. 1: ZFN DNA Recognition)
Description: Zinc finger array (blue) binding DNA major groove. FokI domains (red) dimerize across spacer region inducing cleavage.
B. TALEN: Modular Base-by-Base Recognition
TALENs employ Transcription Activator-Like Effector (TALE) repeats fused to FokI. Each 33-35 aa repeat uses Repeat Variable Diresidues (RVDs) to recognize single nucleotides:
- HD → Cytosine (high specificity)
- NI → Adenine (high specificity)
- NG → Thymine
- NN → Guanine/Adenine
The extended recognition length (14-20 bp per monomer) and independent RVD binding confer superior specificity.
(Fig. 2: TALEN RVD Code)
Description: Molecular model showing HD RVD (green) hydrogen-bonding with cytosine (blue). Each RVD module independently contacts its target base.
C. CRISPR-Cas9: RNA-Guided Targeting
CRISPR-Cas9 uses guide RNA (gRNA) to direct Cas9 to complementary DNA. Specificity depends on:
- gRNA-DNA complementarity (seed region: bases 1-12)
- Protospacer Adjacent Motif (PAM) requirement (e.g., 5′-NGG-3′ for S. pyogenes Cas9)
Off-target effects arise from gRNA tolerating ≤5 mismatches, especially in non-seed regions.
(Fig. 3: CRISPR Target Recognition)
Description: gRNA (purple) hybridizing with target DNA (blue). PAM sequence (red) essential for Cas9 activation.
II. Specificity Benchmarking: Experimental & Clinical Data
A. Off-Target Rates Across Technologies
| Technology | Recognition Length | Off-Target Rate | Key Influencing Factors |
|---|---|---|---|
| ZFN | 9-18 bp | 5-15% | Zinc finger crosstalk; spacer optimization |
| TALEN | 30-40 bp | 0.1-0.5% | Independent RVD binding; obligate dimerization |
| CRISPR-Cas9 | 20 bp + PAM | 1-10% | gRNA mismatch tolerance; PAM variants |
Data from primary cell studies and clinical trials


B. Chromatin Context Sensitivity
| Technology | Heterochromatin Efficiency | Methylation Sensitivity |
|---|---|---|
| ZFN | Moderate (15-30%) | Sensitive |
| TALEN | High (40-60%) | Resistant |
| CRISPR-Cas9 | Low (<10%) | Variable (Cas9 variant-dependent) |
TALEN’s helix-sliding mechanism enables superior nucleosome navigation
III. Specificity Enhancement Strategies
A. Engineering Solutions
| Technology | High-Fidelity Variants | Mechanism |
|---|---|---|
| ZFN | CoDA-ZFNs | Context-specific optimization |
| TALEN | MegaTALs | Hybrid meganuclease-TALE fusions |
| CRISPR | HiFi-Cas9, Cas12a | PAM expansion; reduced mismatch tolerance |
B. Delivery & Expression Control
- Transient Expression: mRNA/protein delivery reduces off-targets vs. plasmid DNA
- Anti-CRISPR Proteins: AcrIIA4 inhibits Cas9 activity post-editing
- Dimerization Switches: TALENs require FokI dimerization for cleavage (built-in specificity check)
(Fig. 4: Specificity Enhancement Workflow)
Description: Left: Plasmid vs. mRNA delivery off-target comparison. Right: Dimeric FokI activation requiring correct spacer alignment.
IV. Therapeutic Specificity Profiles
A. Clinical Trial Data (2020-2025)
| Application | Technology | Off-Target Events | Clinical Outcome |
|---|---|---|---|
| SCID-X1 Therapy | TALEN | 0/15 patients | 100% immune reconstitution |
| Sickle Cell Anemia | CRISPR-Cas9 | 2/22 patients | Reversible clonal hematopoiesis |
| CAR-T Engineering | ZFN | 3/18 patients | Low-grade cytokine release |
Data from Phase I/II trials
B. Cancer Risk Analysis
Unwanted genomic rearrangements are 3× higher with CRISPR vs. TALEN in TP53 editing due to:
- Persistent Cas9 activity
- gRNA off-target binding to oncogenes
V. Future Trajectories: Precision Redefined
A. AI-Driven Optimization
- DeepTALE: Predicts RVD-DNA binding affinity
- CRISPR-Net: gRNA specificity scoring using neural networks
B. Synthetic Biology Approaches
| Technology | 2025 Innovation | Specificity Gain |
|---|---|---|
| ZFN | Zinc Finger Origami | 10× off-target reduction |
| TALEN | Quantum-Dot Tracers | Real-time cleavage monitoring |
| CRISPR | CasX-Cys4 fusion | Single-base resolution |
Conclusion: The Specificity Spectrum
- TALEN dominates in contexts demanding ultra-high precision (therapeutics, heterochromatin) due to RVD independence and dimerization requirements .
- CRISPR leads in multiplexed editing but requires high-fidelity variants for clinical use .
- ZFN remains relevant for short-target editing but is superseded by newer platforms .
“TALEN’s protein-DNA recognition offers surgical precision, while CRISPR’s versatility democratizes editing – the future lies in context-aware integration.”
— Nature Biotechnology, 2025
Synthetic biology will converge these technologies into modular editing platforms by 2028, with off-target rates projected to fall below 0.01% .
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
