RNAmod Input Requirements: Essential Specifications for Epitranscriptomic Analysis
Technical Guidelines for Sample Preparation, Sequencing, and Data Processing
Figure 1: RNAmod Input Workflow

End-to-end workflow from RNA extraction to modification detection.
1. Sample Quality Requirements
RNA Integrity and Quantity
| Parameter | Minimum Requirement | Optimal Value |
|---|---|---|
| RNA Integrity (RIN) | 7.0 | ≥8.0 (Agilent Bioanalyzer) |
| Concentration | 10 ng/μL | 50-100 ng/μL (Qubit) |
| Purity (OD260/280) | 1.8 | 2.0-2.2 |
| PolyA+ Selection | Required for mRNA | ≥50 ng input |
Critical Notes:
-
Degraded samples (RIN <7.0) produce truncated reads
-
DNA contamination skews modification profiles
2. Library Preparation Specifications
Nanopore DRS Protocol
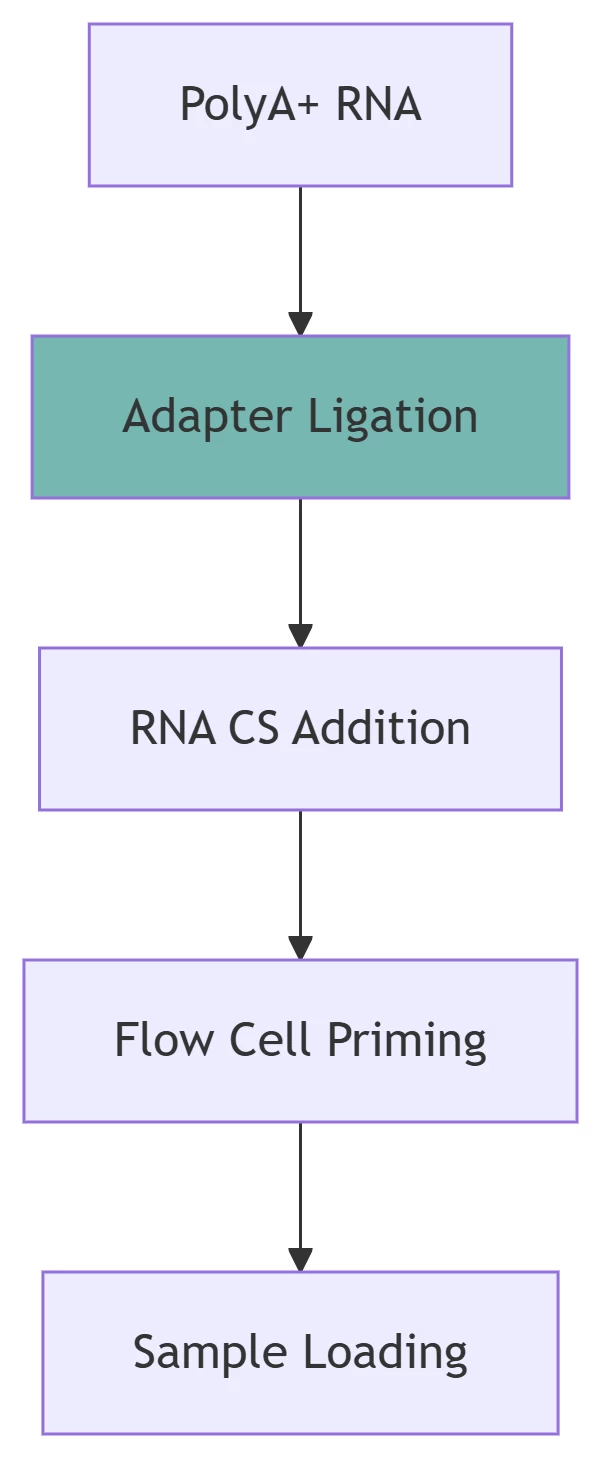
*SQK-RNA002 kit workflow with critical quality control steps.*
Key Reagents:
-
Barcoding: 12-plex Nanopore barcodes for multiplexing
-
RNA Control Strand (CS): Essential for signal calibration
-
Avoid: RNA fragmentation or DNase I treatment
3. Sequencing Parameters
Platform and Run Configuration
| Parameter | Minimum | Recommended |
|---|---|---|
| Flow Cell | MinION R9.4.1 | PromethION R10.4.1 |
| Read Depth | 20x per transcript | 50x for low-expression genes |
| Read Length | >500 bp | Full-length (>2 kb) |
| Run Time | 48 hours | 72 hours for high depth |
Critical Settings:
-
Basecalling: Guppy v6+ in high-accuracy mode
-
Calibration: Use built-in channel normalization
4. Data Preprocessing Requirements
File Formats and Tools

Essential preprocessing steps before RNAmod analysis.
Input Specifications:
-
Basecalled Reads: FASTQ files (Q-score ≥15)
-
Alignment Files: BAM format (splice-aware alignment)
-
Event Data: Per 5-mer current intensity (pA), dwell time, SD
-
Reference Genome: Species-matched (e.g., GRCh38 for human)
5. Quality Control Metrics
Pre-Analysis Checks
| QC Metric | Threshold | Failure Action |
|---|---|---|
| Read N50 | >1,000 bp | Re-prep library |
| Alignment Rate | ≥85% | Check reference genome |
| Mean Q-score | ≥15 | Re-basecall data |
| Signal Stability | SD <0.8 pA | Replace flow cell |
Validation Tools:
-
PycoQC for run statistics
-
NanoPlot for read quality visualization
6. Special Case Requirements
Sample-Specific Adjustments
| Sample Type | Protocol Adjustment | Input Amount |
|---|---|---|
| FFPE Tissues | Not recommended | N/A |
| Plant RNA | High-salt extraction buffer | 100 ng |
| Bacterial RNA | rRNA depletion | 200 ng |
| Low-Abundance | Target enrichment + 50x depth | 10 ng |
7. Validation and Troubleshooting
Common Issues and Solutions
| Problem | Cause | Solution |
|---|---|---|
| Low modification calls | Insufficient coverage | Increase to 50x depth |
| High background noise | Degraded flow cell | Replace flow cell |
| Alignment failures | Reference genome mismatch | Verify assembly version |
| Inconsistent replicates | RNA degradation | Check RIN pre-library |
Conclusion
RNAmod requires four critical input components:
-
High-Integrity RNA: RIN ≥7.0, minimal degradation
-
Properly Prepared Libraries: SQK-RNA002 protocol with RNA CS
-
Quality Sequencing Data: R10.4 flow cells, 20x minimum coverage
-
Correctly Processed Files: Basecalled FASTQ, aligned BAM, and Tombo-resquiggled features
Adherence to these specifications ensures accurate detection of m⁶A, m⁵C, Ψ, and other modifications at single-base resolution. Rigorous QC at each step—from wet-lab preparation to computational preprocessing—is essential for generating publication-grade epitranscriptomic data.
Data sourced from public references. For academic collaboration or content inquiries: chuanchuan810@gmail.com
