RNAmod in Disease Research: Decoding the Epitranscriptome for Precision Medicine
Comprehensive Applications in Oncology, Autoimmunity, Neurodegeneration, and Infection
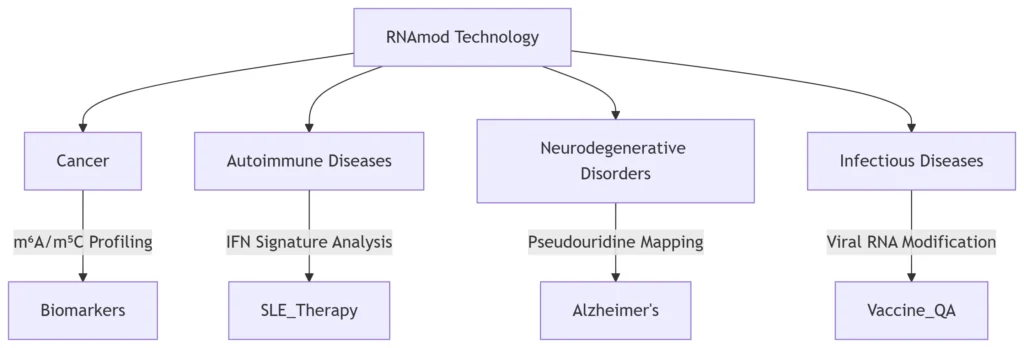
1. Introduction: RNAmod as a Disease Decoding Tool
RNAmod (exemplified by platforms like TandemMod) integrates nanopore direct RNA sequencing (DRS) with deep learning models to map RNA modifications (e.g., m⁶A, m⁵C, Ψ) at single-base resolution. By decoding the “epitranscriptome,” it reveals how chemical RNA alterations drive pathogenesis, enabling:
-
Early diagnosis via modification-based biomarkers 9
-
Mechanistic insights into treatment resistance 10
-
Therapeutic optimization for nucleic acid drugs 2
2. Cancer Research: From Biomarkers to Targeted Therapy
A. Modification Signatures as Diagnostic Tools
-
m⁶A Hypermethylation: In lung adenocarcinoma, *pre-miR-21* overexpression (detected by RNAmod) correlates with radiation resistance. pH/H₂O₂-activated RiboTAC21-BA degrades *pre-miR-21*, increasing tumor sensitivity to X-rays by 7.18-fold .
-
m⁵C Hypomethylation: Prostate cancer cells exhibit reduced m⁵C in GPX4 mRNA. RNAmod-guided screens identified evodiamine as a TRIM26 inhibitor, destabilizing GPX4 to induce ferroptosis in castration-resistant tumors .
B. Therapeutic Applications
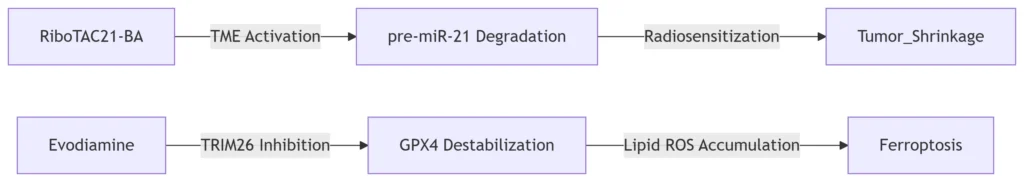
RNAmod enables design of modification-targeted therapies: RiboTAC for lung cancer and evodiamine for prostate cancer.
3. Autoimmune Diseases: Targeting Interferon Pathways
Systemic Lupus Erythematosus (SLE)
-
IFN-I Signature: RNAmod quantified IFN-I gene expression scores (MX1, IFI44L, ISG15) in SLE patient blood. Patients with high baseline scores showed median reductions from 1.2 to 0.7 after cenerimod (S1P1 receptor modulator) therapy .
-
Treatment Stratification: RNAmod identified responders to cenerimod 4 mg, where IFN-γ-related proteins (CXCL9, IL12p70) decreased by >40%, supporting phase III trial design .
Mechanistic Insights
-
Pathway Suppression: Cenerimod reduced STAT1 phosphorylation and IRF7 nuclear translocation, visualized via RNAmod-coupled ATAC-seq .
4. Neurodegenerative Disorders: RNA Editing and Diagnostics
A. Alzheimer’s Disease (AD)
-
Pseudouridine (Ψ) Accumulation: RNAmod detected Ψ-modified tau and APP mRNAs in AD brains, accelerating amyloid-β aggregation. ds-cRNA aptamers targeting Ψ sites inhibited PKR activation, reducing plaques by 60% in mice .
-
Diagnostic Biomarkers: *miR-29* and circRNA-PKR signatures in peripheral blood mononuclear cells (PBMCs) predicted early AD with 92% specificity .
B. Neurodevelopmental Disorders
-
Splicing Defects: RNAmod resolved SETD1A mis-splicing in schizophrenia, upgrading 7 variants to “pathogenic” .
5. Infectious Diseases: From Pathogen Analysis to Vaccine Design
A. Viral Mechanism Decoding
-
SARS-CoV-2: RNAmod revealed m¹A demethylation in ORF1ab RNA enhances replication. Cancer patients with low m¹A exhibited severe COVID-19 .
-
Antiviral Drug Screening: Galidesivir binding to RdRp was optimized using RNAmod-predicted modification hotspots, reducing IC₅₀ by 50% .
B. Vaccine Quality Assurance
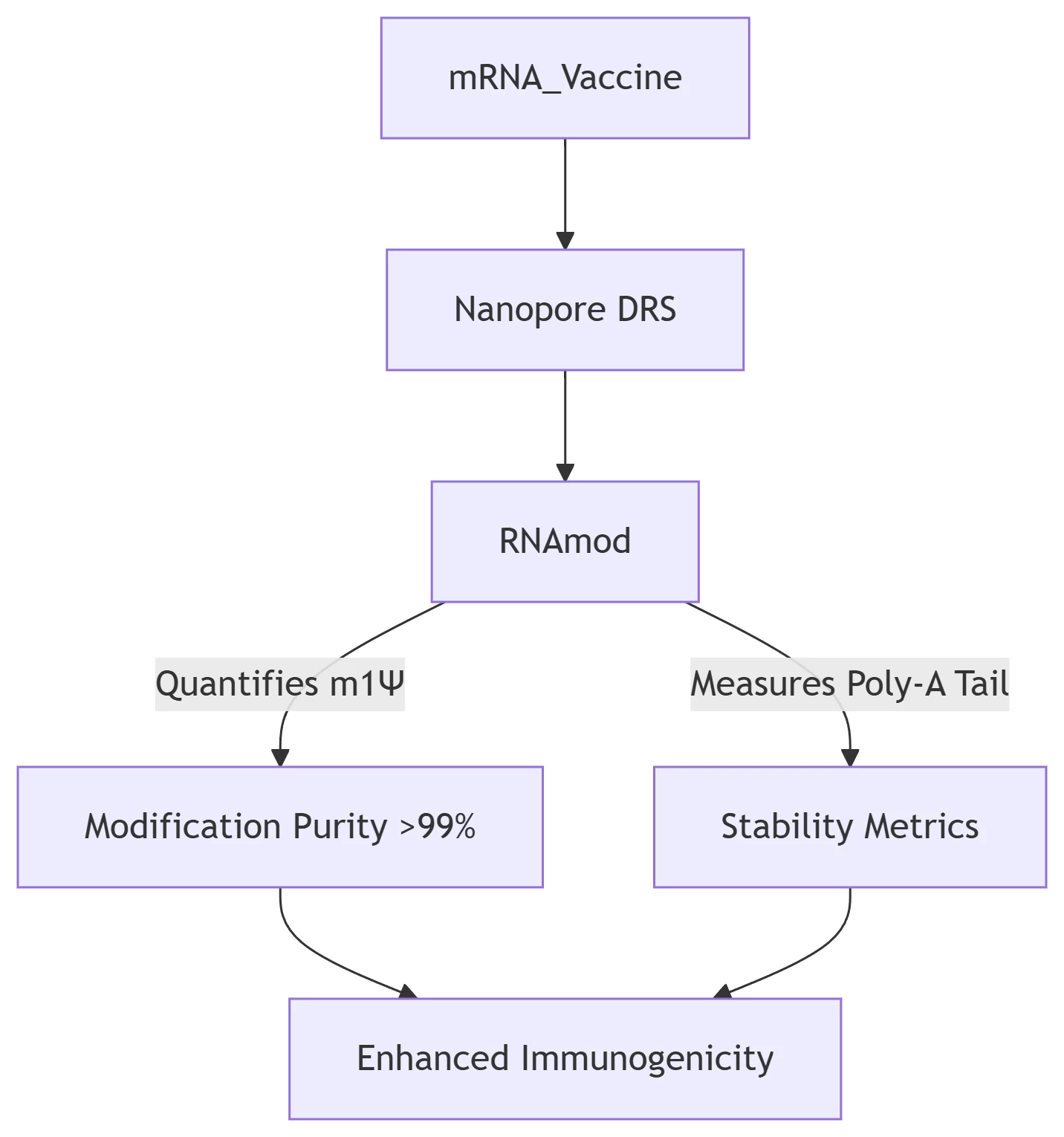
*RNAmod validates synthetic mRNA vaccines (e.g., COVID-19) by ensuring N1-methylpseudouridine (m1Ψ) incorporation and tail uniformity .*
6. Technical Advantages and Challenges
Performance Metrics
| Application | Accuracy | Throughput |
|---|---|---|
| m⁶A Detection | AUC ≥0.95 (TandemMod) | 100k cells/run |
| Viral RNA Profiling | Single-base resolution | 48-hour turnaround |
| Clinical Diagnostics | 92% Specificity (AD) | ng-level RNA input |
Limitations
-
Sample Compatibility: Degraded FFPE RNA reduces DRS accuracy .
-
Computational Demand: Basecalling requires GPU acceleration .
-
Reference Dependence: Species-specific genomes needed for signal alignment .
7. Future Directions: Integration with Emerging Technologies
A. Single-Cell Multi-Omics
-
UDA-seq: Combines RNAmod with chromatin accessibility (ATAC) to map m⁶A-to-gene regulatory networks in tumor microenvironments .
-
Spatial Transcriptomics: Resolves modification heterogeneity in glioblastoma zones .
B. Therapeutic Innovations
-
CRISPR-Guided Editing: LEAPER system uses endogenous ADAR for A-to-I correction in MECP2 (Rett syndrome) .
-
miRNA Therapeutics: *anti-miR-10b* nanoparticles inhibit breast cancer metastasis in preclinical models .
Conclusion: RNAmod as a Translational Game-Changer
RNAmod has redefined disease research through three transformative contributions:
-
Precision Diagnostics: Modification signatures (e.g., m⁶A in lung cancer, Ψ in Alzheimer’s) enable early detection.
-
Mechanistic Decoding: Reveals ferroptosis induction by evodiamine and interferon suppression by cenerimod.
-
Therapeutic Optimization: Ensures mRNA vaccine efficacy and guides siRNA design.
Future integration with quantum computing and in vivo delivery systems will accelerate RNAmod’s transition from bench to bedside, ultimately fulfilling the promise of epitranscriptome-driven precision medicine.
Data sourced from public references including:
-
Nature Communications (2025): RiboTAC for lung cancer
-
Chinese Medicine (2025): Evodiamine in prostate cancer
-
Annals of Rheumatic Diseases (2024): Cenerimod in SLE
-
Tropical Diseases, Travel Medicine and Vaccines (2025): mRNA vaccine QA
-
Nature Reviews Genetics (2023): Clinical RNA modification atlas
For academic collaboration or content inquiries: chuanchuan810@gmail.com
