RNAmod: Core Functions and Technological Innovations in Epitranscriptomics
A Comprehensive Analysis of Multi-RNA Modification Detection
1. Introduction: The Epitranscriptomics Revolution
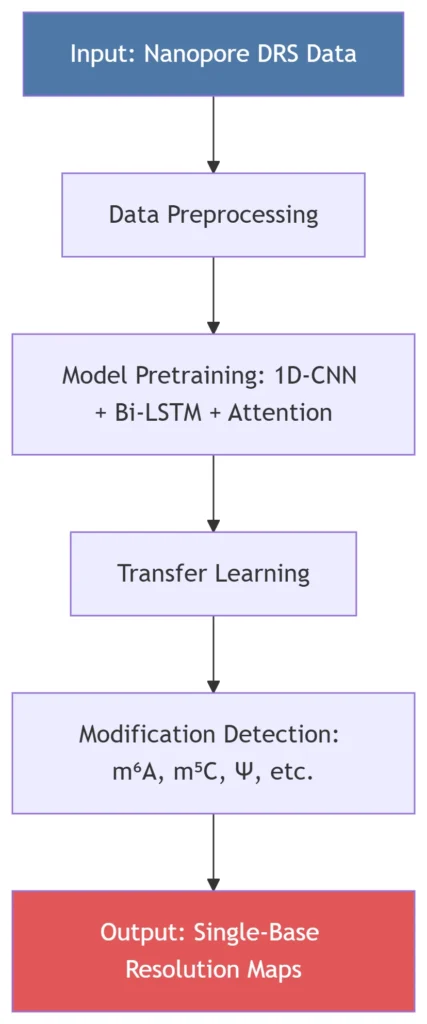
RNA modifications—chemical alterations to RNA nucleotides—regulate critical biological processes, including mRNA stability, translation efficiency, and stress responses. Over 160 natural RNA modifications exist (e.g., m⁶A, m⁵C, Ψ), yet simultaneous detection of multiple modifications at single-base resolution has remained challenging35. RNAmod (exemplified by tools like TandemMod) addresses this gap by integrating nanopore direct RNA sequencing (DRS) with deep learning to decode the “epitranscriptome” with unprecedented accuracy47.
2. Core Functional Capabilities
A. Multi-Modification Detection in Single Molecules
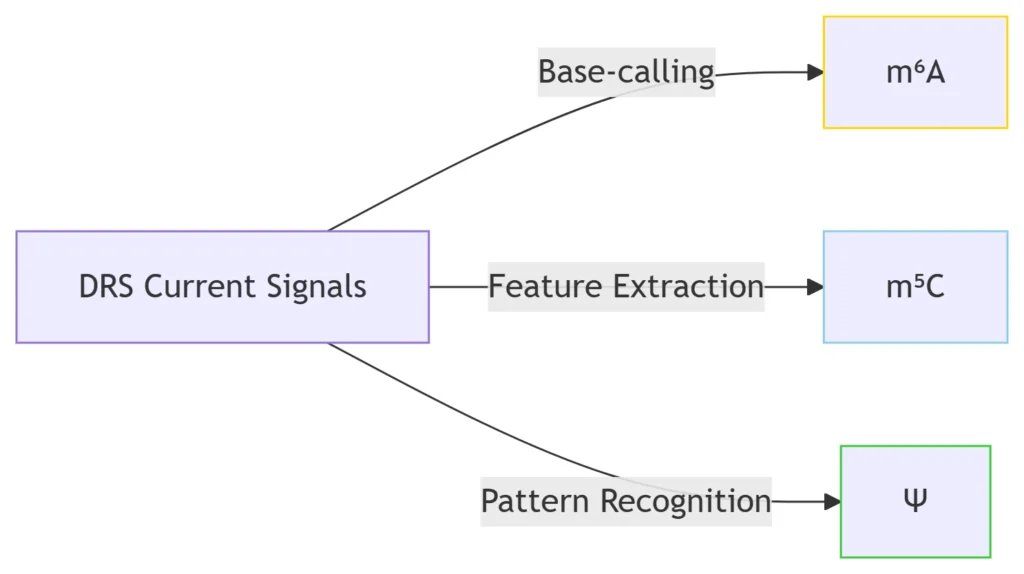
RNAmod simultaneously identifies >6 RNA modification types from one DRS dataset:
-
Common modifications: m⁶A (N⁶-methyladenosine), m⁵C (5-methylcytosine), Ψ (pseudouridine)47
-
Rare modifications: hm⁵C (5-hydroxymethylcytosine), m⁷G (7-methylguanosine), Inosine5
-
Detection accuracy: ROC-AUC ≥0.95 for m⁶A, m⁵C, and Ψ in human and plant transcripts57
B. Deep Learning Architecture
The TandemMod framework leverages:
-
1D Convolutional Neural Networks (1D-CNN): Extracts local features from raw current signals57
-
Bidirectional LSTM (Bi-LSTM): Captures long-range dependencies in RNA sequences7
-
Attention Mechanism: Weights critical signal regions for high-precision modification calling5
C. Transfer Learning for Cost Efficiency
RNAmod reduces computational resources by >50% through:
-
Pretraining: On in vitro epitranscriptome (IVET) datasets (e.g., rice cDNA libraries with m⁶A/m⁵C labels)34
-
Fine-tuning: Freezing top layers and retraining classifiers for new modifications (e.g., m⁷G, Inosine) with minimal data57
3. Functional Workflow & Validation
Step 1: Data Generation
-
IVET Datasets: Synthetic RNA transcripts with known modifications (e.g., m⁶A-tagged mRNAs) via T7 polymerase4
-
In vivo DRS: Native RNA from human cells or stressed plants (e.g., salt-treated rice seedlings)37
Step 2: Model Training & Benchmarking
Metric TandemMod Traditional Tools (Tombo/xPore) m⁶A AUC (DRACH) 0.95 0.71 (m6Anet) Training Time 2 epochs >10 epochs Data Required 40% less Full datasets Performance validated on ELIGOS and Curlcake datasets57 Step 3: Biological Applications
-
Co-modification Mapping: Revealed m⁶A and m⁵C co-occurrence in salt-stressed rice mRNA47
-
Clinical Diagnostics: Detects aberrant m⁶A in METTL3-knockout human cells (linked to cancer)5
-
Vaccine QA: Validates chemical modifications in synthetic mRNA vaccines47
4. Technological Impact
A. Epitranscriptome Atlas Construction
RNAmod generates tissue- and condition-specific modification maps:
-
Case Study: High-resolution m⁶A/m⁵C/Ψ profiles in rice under salt stress showed:
-
m⁶A ↑ 1.8-fold in stress-response genes
-
m⁵C ↓ 40% in metabolic pathways45
-
B. Integration with Disease Research
-
COVID-19: m¹A demethylation enhances SARS-CoV-2 replication, explaining severe symptoms in cancer patients1
-
Genetic Disorders: RNAmod detects splicing-regulatory modifications missed by DNA sequencing (e.g., in BRCA1 VUS)10
5. Advantages Over Existing Methods
Feature RNAmod/TandemMod Conventional RNA-Seq Modification Types Simultaneous multi-target Single-modification focus Resolution Single-base, full-length Indirect inference (antibody/IP) Throughput 10× faster post-training Weeks for library prep Tissue Requirements Low-input (ng RNA) μg-level input
Conclusion
RNAmod represents a paradigm shift in epitranscriptomics by:
-
Democratizing Multi-Mod Detection: Single-assay profiling of >6 modifications
-
Reducing Computational Barriers: Transfer learning slashes data/training needs
-
Enabling Precision Biology: Maps condition-specific epitranscriptomes (e.g., stress, disease)
Future iterations will target in situ modification tracking and direct clinical diagnostics. As the “epitranscriptomic code” is deciphered, RNAmod will accelerate therapies targeting RNA modifications in cancer, viral infections, and genetic disorders.
Data sourced from public references including:
-
Yuan et al., Nature Communications (2024): TandemMod methodology47
-
Fang & Yuan, ChemBioChem (2023): RNA modifications in SARS-CoV-21
-
Genetics in Medicine Open (2025): Clinical RNA-seq integration10
-
RCSB PDB & ELIGOS datasets: Structural and benchmark data
For academic collaboration or content inquiries: chuanchuan810@gmail.com
-
