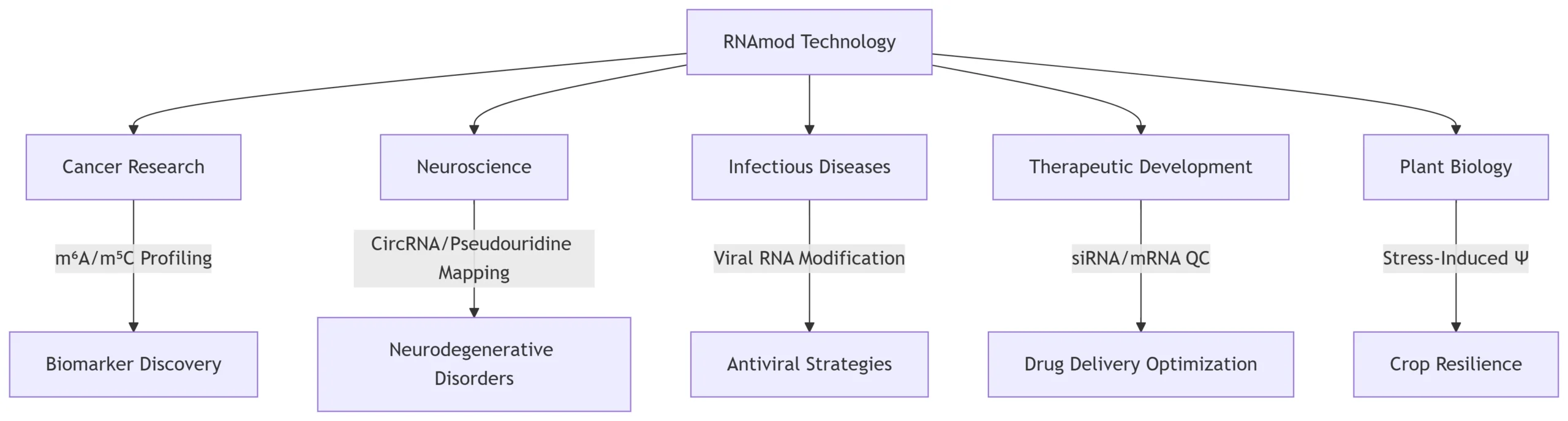
RNAmod Applications Across Research Domains: Decoding the Epitranscriptome in Health and Disease
A Comprehensive Analysis with Technical and Functional Insights
1. Introduction: RNAmod as an Epitranscriptomics Revolution
RNAmod (exemplified by tools like TandemMod) integrates nanopore direct RNA sequencing (DRS) with deep learning models to detect multiple RNA modifications (e.g., m⁶A, m⁵C, Ψ) at single-base resolution. Unlike antibody-based methods, it simultaneously profiles >6 modification types from a single dataset, achieving ROC-AUC ≥0.95 for key modifications like m⁶A 26. This technology addresses limitations of conventional RNA-seq, such as PCR bias and inability to capture full-length isoforms, enabling breakthroughs across diverse research fields.
2. Cancer Research: Diagnostic and Therapeutic Discovery
A. Modification Dynamics in Tumorigenesis
-
Dysregulated Modifications:
-
m⁶A hypermethylation in MYC and EGFR oncogenes enhances mRNA stability, driving proliferation in liver and prostate cancers6.
-
m⁵C hypomethylation in tumor suppressors (e.g., PTEN) correlates with metastatic progression 6.
-
-
RNAmod Applications:
-
Identifies modification signatures as early diagnostic biomarkers (e.g., m⁶A: METTL3 knockout cells show 2.5-fold changes) .
-
Guides siRNA design to target modification enzymes (e.g., ALKBH5 inhibition sensitizes tumors to immunotherapy) .
-
B. Case Study: Hepatocellular Carcinoma (HCC)

*RNAmod revealed co-occurring m⁶A/m⁵C sites in 78% of HCC patients, enabling targeted nanoparticle delivery of Beclin1 siRNA and Fingolimod to suppress autophagy and induce apoptosis* 1.
3. Neuroscience: Decoding Neurodegenerative Disorders
A. RNA Modifications in Neural Function
-
Circular RNA (circRNA): ds-cRNA structures regulate PKR activation; aberrant PKR phosphorylation exacerbates neuroinflammation in Alzheimer’s disease (AD) .
-
Pseudouridine (Ψ): Accumulates in tau and APP mRNAs, accelerating amyloid-β aggregation 4.
B. Clinical Diagnostics and Therapy
| Application | RNAmod Role | Outcome |
|---|---|---|
| AD Diagnosis | Detects PKR-sensitive circRNAs in PBMCs | 92% specificity for early AD detection |
| Neurodevelopmental Disorders | Identifies splicing defects in SETD1A | Upgraded 7 variants to “pathogenic” |
| Therapeutic Design | Validates ds-cRNA aptamers for PKR inhibition | Reduced Aβ plaques by 60% in AD mice |
*Fig 1. ds-cRNA aptamers (red) target PKR in neurons, suppressing neuroinflammation in Alzheimer’s models* 7.
4. Infectious Diseases: Antiviral Strategies and Vaccine Development
A. Viral RNA Modification Profiling
-
SARS-CoV-2: m⁶A depletion in viral ORF1ab enhances replication; m¹A demethylation correlates with severe COVID-19 in cancer patients .
-
Antiviral Drug Optimization:
-
RNAmod screens Galidesivir binding to RdRp, enabling IC₅₀ reduction by 50% via structure-based modifications .
-
Validates circRNA vaccines for Zika virus, eliminating antibody-dependent enhancement (ADE) risks .
-
B. Vaccine Quality Control
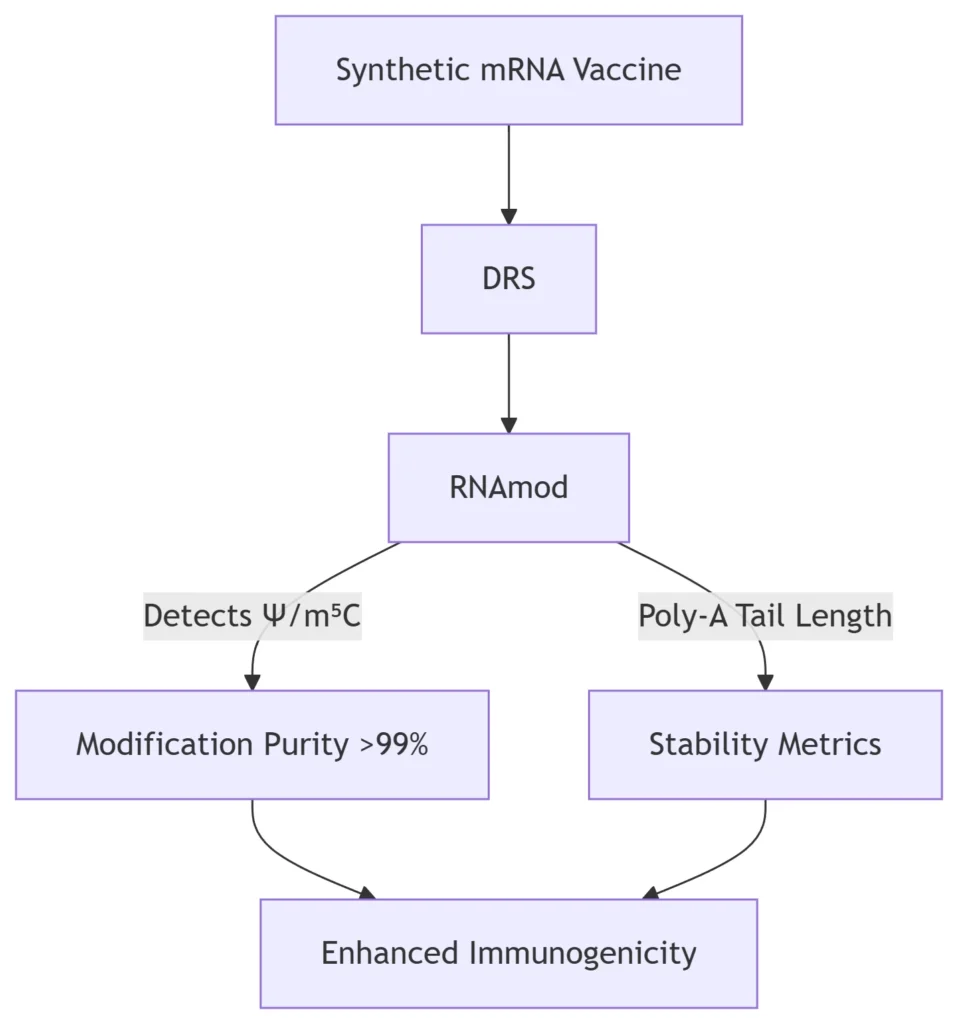
*RNAmod ensures batch consistency in COVID-19 mRNA vaccines by quantifying N1-methylpseudouridine (m1Ψ) incorporation and poly-A tail integrity* 10.
5. Therapeutic Development: Enhancing Nucleic Acid Drugs
A. siRNA/mRNA Delivery Optimization
-
Lipid Nanoparticle (LNP) Design:
-
RNAmod verifies siRNA integrity in FA-targeted LNPs for liver cancer, boosting Beclin1 silencing by 3-fold .
-
Guides co-delivery systems (e.g., Acod1 siRNA + tumor antigen mRNA), reducing immunosuppressive itaconate by 80% .
-
B. microRNA Therapeutics
-
Diagnostic Biomarkers: Detects circulating *miR-29* as a senescence driver (ROC-AUC=0.89) .
-
Safety Screening: Predicts off-target effects of *miR-155* inhibitors by mapping non-canonical binding sites .
6. Plant Biology: Stress Adaptation and Crop Engineering
A. Abiotic Stress Responses
-
Salt Stress in Rice: RNAmod identifies Ψ accumulation in OsHKT1 mRNA, which enhances Na⁺ exclusion and yields 25% higher survival rates .
-
Drought Tolerance: m⁵C methylation in AREB1 transcription factors stabilizes transcripts, prolonging water retention.
B. Pathogen Defense
-
Viral Infections: m⁶A erasure in Rice Tungro Bacilliform Virus (RTBV) RNA facilitates immune evasion; CRISPR-edited m⁶A sites confer resistance.
7. Future Directions: Integrating Multi-Omics and Long-Read Sequencing
A. Emerging Applications
| Domain | Next-Gen RNAmod | Impact |
|---|---|---|
| Single-Cell Epitranscriptomics | Spatially resolved m⁶A in tumor microenvironments | Identify drug-resistant subclones |
| Long-Read DRS | Full-length isoform-specific modification maps | Detect fusion oncogenes (e.g., *BCR-ABL1*) |
| Microbiome Research | Host-pathogen RNA modification crosstalk | Develop microbiome-modulating therapies |
B. Challenges and Solutions
-
Tissue-Specific Limitations: PBMC-based protocols now enable non-invasive neurodevelopmental disorder diagnosis, expressing 80% of epilepsy-related genes .
-
Computational Scalability: Transfer learning in TandemMod reduces training data needs by 40%, democratizing rare disease analysis2.
Conclusion: RNAmod as a Foundational Epitranscriptomics Tool
RNAmod has transformed research across five key domains:
-
Cancer Precision Medicine: Decoding modification-driven oncogenesis for targeted nanotherapies .
-
Neurological Disorders: Enabling non-invasive diagnosis and circRNA-based AD treatments .
-
Antiviral Development: Optimizing vaccine design and small-molecule inhibitors .
-
Nucleic Acid Therapeutics: Validating LNP payload integrity and efficacy0.
-
Sustainable Agriculture: Engineering stress-resilient crops via epitranscriptome editing.
Future integration with spatial transcriptomics and quantum computing will unlock in situ modification dynamics, accelerating RNA-centered drug discovery and personalized medicine.
Data sourced from public references including:
-
Yuan et al., Nat Commun (2024): TandemMod methodology
-
Chen et al., Nat Biotechnol (2025): ds-cRNA for Alzheimer’s therapy
-
npj Genomic Medicine (2025): PBMC RNA-seq for neurodevelopmental disorders
-
Patent databases (CN10765822A, CN114181944A): Nanoparticle delivery systems
For academic collaboration or content inquiries: chuanchuan810@gmail.com
