RNAmod: Advanced Methodologies for Programmable RNA Editing
A Comprehensive Analysis of Molecular Tools, Delivery Strategies, and Therapeutic Applications
1. Introduction: RNA Editing as a Therapeutic Paradigm
RNA editing technologies enable site-specific modifications of RNA transcripts without altering genomic DNA, offering reversible and controllable correction of disease-causing mutations. Key advantages over DNA editing include:
-
Reversibility: Transient edits reduce off-target risks
-
Spatiotemporal control: Light-inducible systems enable precise activation
-
Avoidance of double-strand breaks: Eliminates genomic instability
RNAmod integrates these features into a unified framework for research and therapy.
2. Core Methodologies in RNAmod
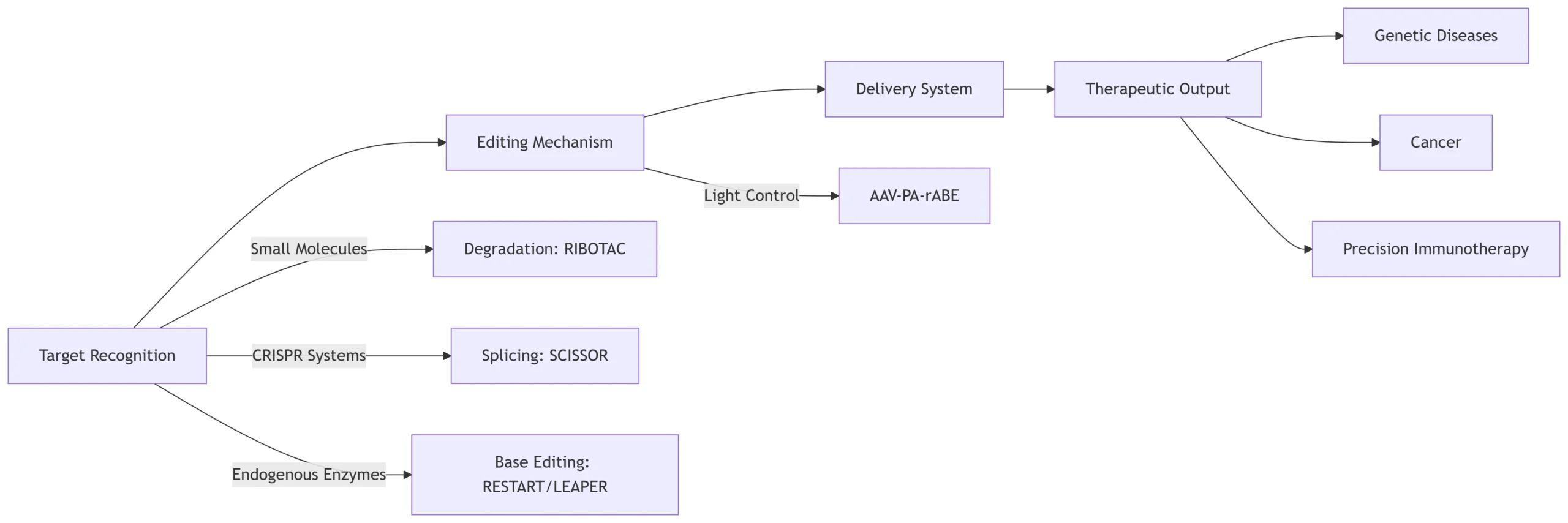
A. Target Recognition Strategies
1. Small Molecule-Based Recognition (RIBOTAC)
-
Mechanism: Inactive RNA-binding molecules are linked to RNase L recruiters, enabling targeted degradation of structured RNAs .
-
Example: Degradation of oncogenic MYC mRNA in B-cell lymphoma (efficiency: >90%)

2. CRISPR-Derived Systems (SCISSOR)
-
Innovation: Engineered bulge loops in III型 CRISPR-Csm enable arbitrary-length RNA deletions .
-
Application: Frameshift correction in HEXA gene for Tay-Sachs disease (80% PTC readthrough) .
3. Endogenous Enzyme Recruitment (RESTART/LEAPER)
-
RESTART: Guide snoRNAs direct pseudouridine synthase to convert stop codons (UAA/UAG) to ΨAA/ΨAG for PTC readthrough .
-
LEAPER: Single arRNA recruits endogenous ADAR for A-to-I editing (efficiency: ≤80%).
B. Editing Mechanisms
Mechanism Tool Edit Type Precision Base Editing PA-rABE A-to-I (light-controlled) 95% spatial specificity Exon Reframing SCISSOR Arbitrary deletion 2-bp resolution RNA Degradation RIBOTAC Site-specific cleavage RNase L-dependent Pseudouridylation RESTART U-to-Ψ No codon alteration C. Delivery Systems
1. Viral Vectors
-
AAV: Used for in vivo delivery of RESTART (snoRNA) and PA-rABE (split ADAR) .
-
Lentivirus: Stable expression of LEAPER arRNAs in T cells .
2. Non-Viral Platforms
-
GalNAc-conjugated snoRNAs: Liver-targeted RESTART delivery .
-
LNP-encapsulated RIBOTACs: Tumor-specific MYC degradation .
3. Activable Nanocarriers
-
Light-responsive AAVs: Spatiotemporal control of PA-rABE in hemophilia B mice .
3. Therapeutic Applications
A. Genetic Disease Correction
1. Premature Termination Codon (PTC) Readthrough
-
RESTART: Corrected IDUA deficiency in Hurler syndrome cells via Ψ-mediated PTC suppression.
-
Efficiency: >60% functional protein restoration in bronchial epithelial cells .
2. Neurological Disorders
-
LEAPER: Repaired MECP2 nonsense mutations in Rett syndrome models using endogenous ADAR .
-
Safety: No immune response (human-derived components) .
B. Cancer Therapy
1. Oncogene Targeting
-
RIBOTAC: Degraded JUN/MYC mRNAs in breast cancer and lymphoma (IC₅₀: 0.5 μM) .
-
SCISSOR: Induced frameshifts in HER2, generating immunogenic neoantigens .
2. Combination Immunotherapy
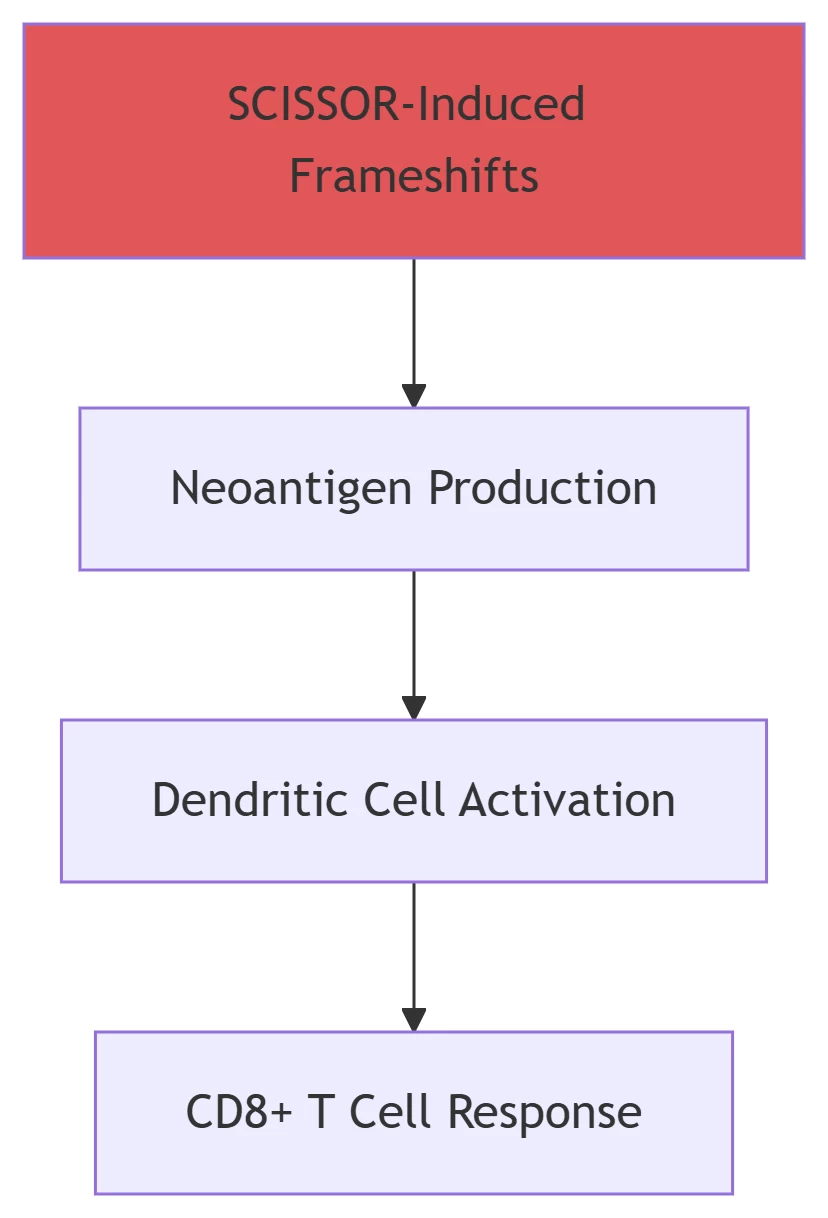
Frameshifted oncoproteins activate antitumor immunity .
C. Precision-Controlled Editing
PA-rABE System in Hemophilia B:
-
Components:
-
mini dCas13X: RNA-targeting module
-
Split ADAR2dd: Light-activatable deaminase
-
-
Workflow:
-
Blue light → pMag/nMag dimerization → ADAR2dd reconstitution → F9 mRNA repair
-
-
Outcome:
-
Coagulation factor IX restored to 40% normal levels .
-
4. Methodological Advantages & Limitations
Performance Comparison
Tool Editing Window Off-Target Rate Key Advantage LEAPER Flexible 0.1% No exogenous proteins SCISSOR 5-100 nt Undetectable Frameshift correction PA-rABE 1-2 nt <0.05% Spatiotemporal control RIBOTAC Structural pockets RNase L-dependent Degrades “undruggable” RNAs Technical Challenges
-
Delivery Efficiency:
-
AAV cargo limit (<4.7 kb) restricts SCISSOR/PA-rABE packaging.
-
-
Endogenous Competition:
-
LEAPER arRNAs outcomped by cellular RNAs reduce editing efficiency .
-
-
Immune Activation:
-
RIBOTACs may trigger IFN responses via RNase L activation .
-
5. Future Directions
A. Integration with Multi-Omics
-
RNAmod-Atlas: Nanopore DRS maps m⁶A/Ψ modifications to predict editable sites .
-
AI-Guided Design:
-
AlphaFold-RNA: Predicts targetable RNA folds for RIBOTAC development .
-
B. Clinical Translation
-
Ex Vivo Cell Therapy:
-
CAR-T cells with PA-rABE-controlled PD1 knockout .
-
-
In Vivo Applications:
-
GalNAc-RESTART for liver diseases (clinical trials by 2026) .
-
C. Synthetic Biology
-
RNA-Binding Protein Switches:
-
Fusion of RIBOTAC recruiters to aptamers for biomarker-responsive editing .
-
Conclusion
RNAmod represents a paradigm shift in RNA-targeted therapeutics through three key innovations:
-
Precision Recognition: Small molecules and CRISPR systems enable targeting of structured RNA domains.
-
Diverse Editing Outcomes: Base editing, exon reframing, and degradation address varied mutation types.
-
Controllable Delivery: Light-inducible and tissue-specific systems minimize off-target effects.
The integration of these methodologies—exemplified by RIBOTAC-driven oncogene degradation 1, RESTART-mediated PTC suppression 2, and PA-rABE’s spatiotemporal control 8—will accelerate treatments for genetic diseases, cancer, and regenerative disorders. Future advances in in vivo delivery and computational design will further establish RNAmod as the cornerstone of next-generation gene therapy.
Data sourced from public references including:
-
Disney Lab, Nature (2023): RIBOTAC technology
-
Yi Lab, Molecular Cell (2022): RESTART system
-
Zhang Lab, Molecular Cell (2025): SCISSOR applications
-
Wei Lab, Nature Biotechnology (2019): LEAPER efficiency
-
Li Lab, Nature Biotechnology (2025): PA-rABE characterization
For academic collaboration or content inquiries: chuanchuan810@gmail.com
-
