A Step-by-Step Analysis from Initiation to Termination

1. Overview of Transcription
Transcription is the process by which genetic information encoded in DNA is copied into RNA by RNA polymerase (RNAP). This RNA may serve as:
-
Messenger RNA (mRNA) for protein synthesis
-
Functional RNA (tRNA, rRNA, regulatory RNAs)
The process occurs in three phases: initiation, elongation, and termination, differing in prokaryotes vs. eukaryotes.
2. Transcription Initiation
A. Promoter Recognition
-
Core Promoter Elements:
-
TATA box (eukaryotes): Recognized by TATA-binding protein (TBP) subunit of TFIID
-
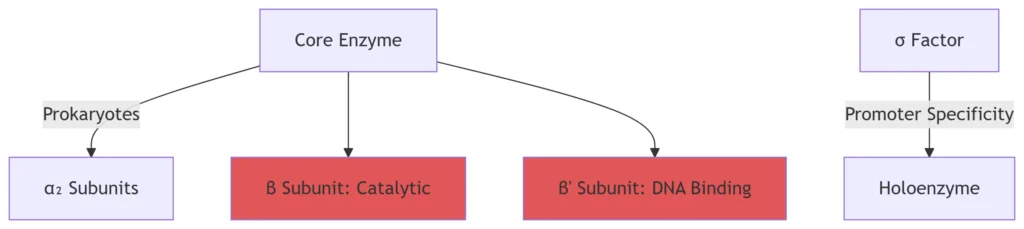
-10 box (prokaryotes): Consensus sequence “TATAAT” bound by σ factor
-
-
Transcription Factors:
-
Eukaryotes require 6 GTFs (TFIID, TFIIA, TFIIB, TFIIF, TFIIE, TFIIH)
-
Prokaryotes use σ factors (e.g., σ⁷⁰) for promoter specificity
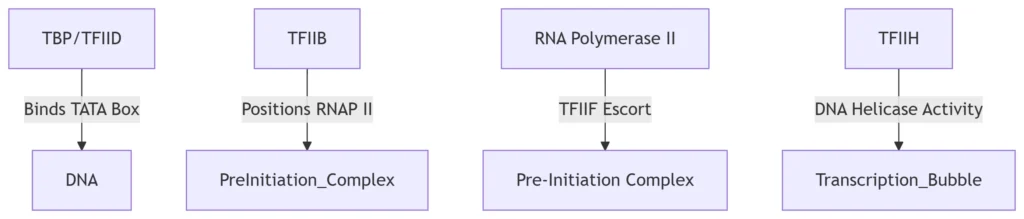
PIC assembly bends DNA 80°, creating a scaffold for RNAP II recruitment.
B. Open Complex Formation
-
DNA Unwinding:
-
TFIIH hydrolyzes ATP to unwind DNA (14-bp transcription bubble)
-
Template strand enters RNAP active site
-
3. Elongation Phase
A. RNA Synthesis Mechanics
-
Directionality:
-
RNA synthesized 5’→3′
-
Template DNA read 3’→5′
-
-
Catalytic Mechanism:
-
Nucleotides added via phosphodiester bond formation
-
Error rate: 1/10⁴ bases (no proofreading)
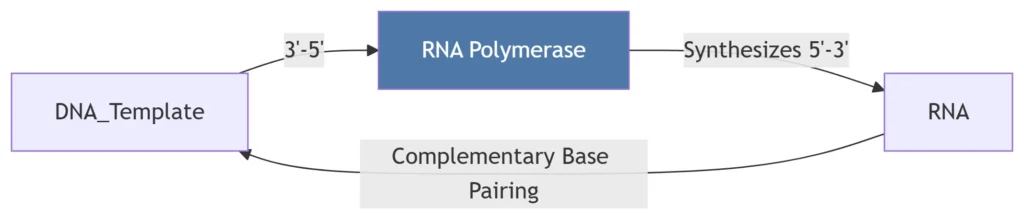
*RNA-DNA hybrid (8-9 bp) maintained in active site.*
B. Elongation Complex Dynamics
Component Function RNAP Active Site Catalyzes nucleotide addition Downstream DNA Unwound ahead of polymerase Upstream DNA Rewound behind polymerase Transcript Exit Tunnel Channels nascent RNA away
4. Transcription Termination
A. Prokaryotic Termination
-
Rho-Dependent:
-
Rho helicase binds rut site on RNA, translocates to unwind RNA-DNA hybrid
-
-
Rho-Independent:
-
GC-rich hairpin + poly-U sequence stalls RNAP
-
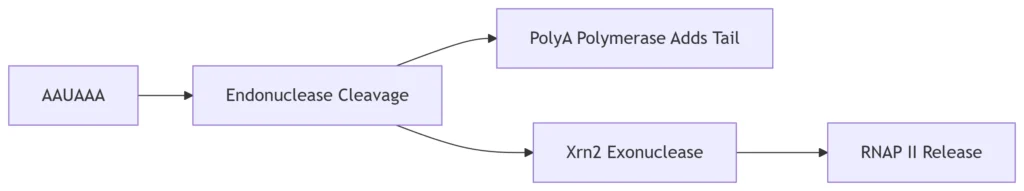
B. Eukaryotic Termination
-
Polyadenylation Signal (AAUAAA):
-
Triggers RNA cleavage 10-30 nt downstream
-
-
Torpedo Model:
-
Xrn2 exonuclease degrades transcript, dislodges RNAP II
5. Post-Transcriptional Processing (Eukaryotes)
Modification Function Molecular Mechanism 5′ Capping Protects mRNA, aids translation 7-methylguanosine added co-transcriptionally Splicing Removes introns Spliceosome recognizes GU-AG boundaries 3′ Polyadenylation Stabilizes mRNA, enables export 200-250 adenine tail added 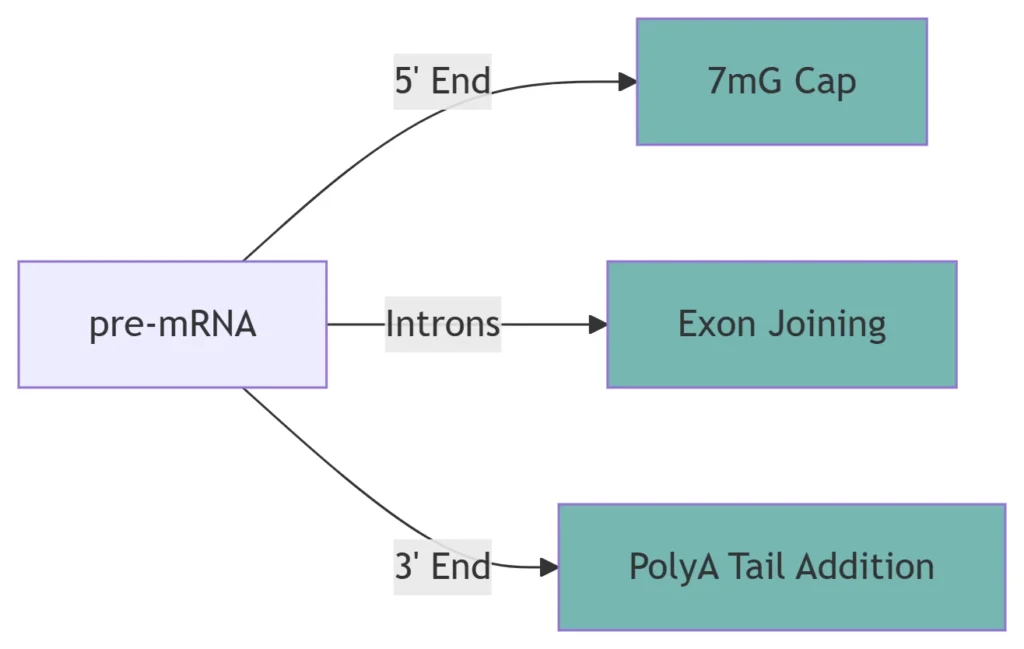
6. Key Structural Insights
RNA Polymerase Architecture
Eukaryotic RNAP II contains 12 subunits, with RPB1 forming the active site.
Conclusion
Transcription converts DNA to RNA through three stages:
-
Initiation: Promoter recognition and PIC assembly
-
Elongation: Processive RNA synthesis (5’→3′)
-
Termination: Signal-dependent polymerase release
Eukaryotic transcripts undergo extensive processing (capping, splicing, polyadenylation) to generate functional mRNAs. This highly conserved process occurs at 40-80 nucleotides/second, balancing speed and accuracy across all domains of life.
Data sourced from public references. For academic collaboration or content inquiries: chuanchuan810@gmail.com
-
-
-
-
-
-
