A Comprehensive Analysis of Transcriptional Control
Figure 1: Transcriptional Regulation Workflow

1. Transcriptional Initiation: The Primary Control Point
A. Chromatin Accessibility
-
Histone Modifications:
-
H3K27ac → Open chromatin → 80% of active promoters
-
H3K27me3 → Closed chromatin → gene silencing
-
-
Chromatin Remodelers:
-
SWI/SNF complexes slide nucleosomes away from promoters
-
B. Pre-Initiation Complex (PIC) Assembly
Key Components:
| Factor | Function | Regulatory Mechanism |
|---|---|---|
| TFIID | TATA-box recognition | Phosphorylation by CDK7 |
| Mediator | RNAP II recruitment | Signal-dependent conformation |
| TFIIH | DNA unwinding | Kinase activity regulation |
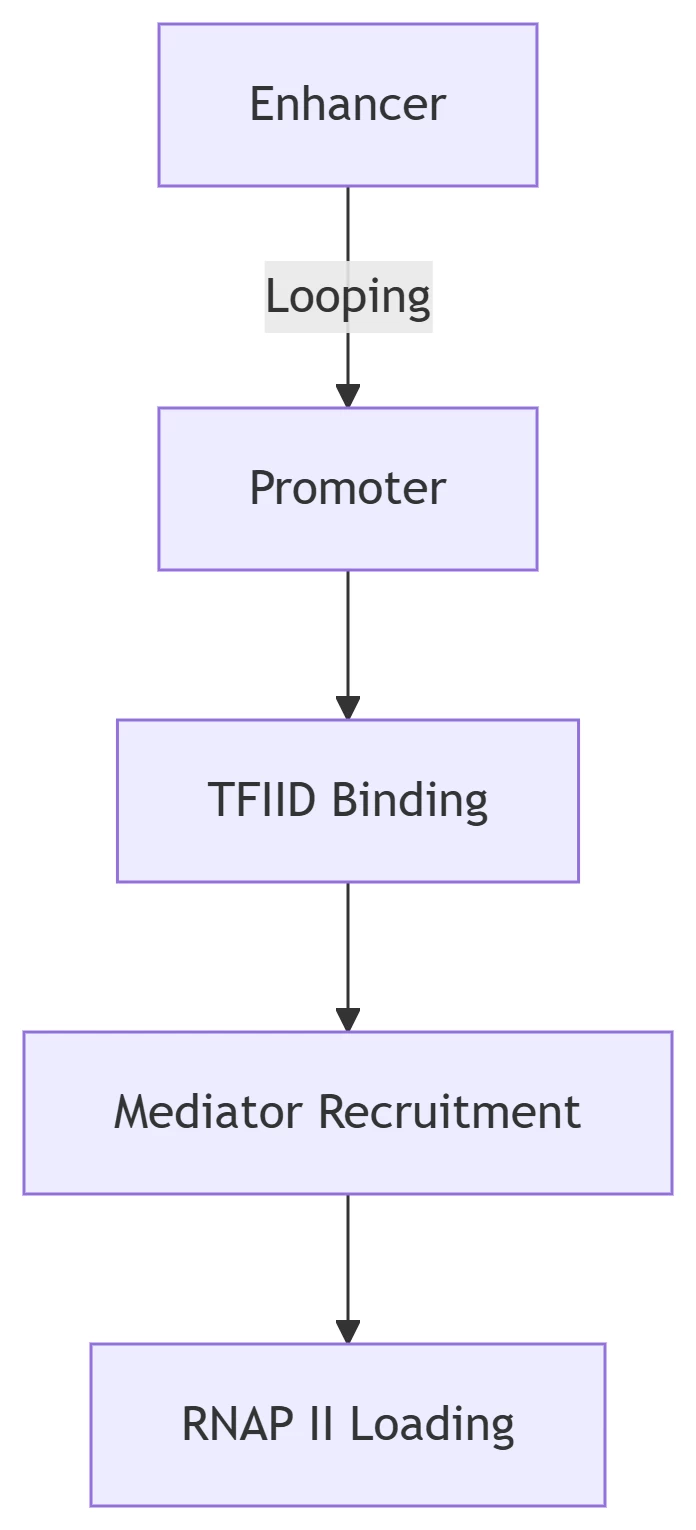
2. Paused Polymerase: The Regulatory Checkpoint
A. RNAP II Pausing Mechanisms
-
DSIF/NELF Complex:
-
Binds nascent RNA to stall RNAP II 30-60 bp downstream
-
-
P-TEFb Activation:
-
Phosphorylates DSIF/NELF → pause release
-
-
Biological Functions:
-
Allows rapid response to stimuli (e.g., heat shock)
-
Synchronizes transcription with splicing
-
B. Pause Release Triggers
| Signal | Release Factor | Target Gene Example |
|---|---|---|
| Growth Factors | MYC | CCND1 (Cyclin D1) |
| Stress | HSF1 | HSP70 |
| Immune Activation | NF-κB | IL6 |
3. Elongation Control: Beyond Initiation
A. Co-Transcriptional Processing
-
Splicing Coupling:
-
RNAP II CTD coordinates spliceosome assembly
-
Speed-dependent alternative splicing outcomes
-
-
mRNA Modification:
-
m⁶A deposition by METTL3 during elongation
-
B. RNAP II Speed Modulation
Speed Regulators:
| Factor | Effect | Biological Consequence |
|---|---|---|
| TFIIS | ↑ Speed | Error correction |
| H2Bub1 | ↓ Speed | Splicing fidelity |
| PAF1 Complex | ↑ Speed | Enhancer RNA production |
4. Termination and Feedback Regulation
A. Termination-Linked Mechanisms
-
PolyA Signal Recognition:
-
CPSF complex binds AAUAAA → cleavage
-
-
Termination Consequences:
-
Xrn2 “torpedo” degrades transcript
-
RNAP II recycling for new initiation
-
B. Transcriptional Interference
-
Antisense Transcription:
-
Overlapping genes block elongation
-
-
Regulatory Function:
-
Controls imprinted genes (e.g., IGF2-H19 locus)
-
5. Regulatory Layer Integration
Hierarchical Control Points:
| Layer | Key Mechanism | Response Time |
|---|---|---|
| Chromatin | Histone modifications | Hours-days |
| PIC Assembly | TF recruitment dynamics | Minutes |
| Pause Release | P-TEFb activation | Seconds-minutes |
| Elongation Speed | RNAP II phosphorylation | Real-time |
6. Disease Relevance
A. Cancer-Linked Dysregulation
| Gene | Regulatory Defect | Cancer Type |
|---|---|---|
| MYC | Pause release hyperactivation | Lymphoma, Breast |
| BRCA1 | Promoter methylation | Ovarian, Breast |
| AR | Enhancer hijacking | Prostate |
B. Therapeutic Targeting
-
CDK9 Inhibitors (e.g., Flavopiridol): Block pause release
-
BET Inhibitors (e.g., JQ1): Prevent enhancer factor binding
7. Technological Advances
Single-Molecule Imaging
-
Live-Cell RNAP Tracking:
-
MS2-GFP system reveals elongation kinetics
-
Identified 7 distinct RNAP II states
-
-
Chromatin Conformation Capture:
-
Hi-C maps promoter-enhancer interactions
-
Conclusion
RNA transcription regulates gene expression through three hierarchical control points:
-
Initiation: Chromatin accessibility and PIC assembly determine transcriptional competence
-
Pause Release: Serves as rapid response checkpoint for signals
-
Elongation: Speed modulation coordinates RNA processing
These mechanisms enable precise spatiotemporal control—from milliseconds (elongation speed) to days (chromatin remodeling)—with dysregulation causing cancer, neurodegeneration, and developmental disorders. Emerging single-molecule technologies continue to decode transcriptional dynamics at unprecedented resolution.
Data sourced from public references. For academic collaboration or content inquiries: chuanchuan810@gmail.com
