 I. Foundational Classification Framework
I. Foundational Classification Framework
RNA probes constitute a specialized class of nucleic acid detectors engineered for sequence-specific hybridization. Three primary structural archetypes dominate molecular diagnostics and research:
A. cRNA Probes (In Vitro Transcribed)
- Source: DNA templates with phage promoters (T7/T3/SP6)
- Structure: Single-stranded RNA (ssRNA) with full-length complementarity
- Production Workflow:

- (Fig. 1: In vitro transcription schematic)
Description: Molecular visualization of RNA polymerase binding promoter sequence and synthesizing labeled cRNA.
B. cDNA Probes (Reverse-Transcribed)
- Source: mRNA templates via reverse transcriptase
- Structure: Complementary DNA strands with optional ssRNA regions
- Key Applications:
- Low-abundance transcript detection
- Mutation-specific diagnostics (e.g., BRAF V600E)
C. Oligonucleotide Probes (Chemically Synthesized)
- Source: Solid-phase phosphoramidite synthesis
- Structure: Short ssRNA (12-50 nt) with terminal modifications
- Design Innovations:
- Locked Nucleic Acids (LNA) for enhanced stability
- Molecular beacons with stem-loop quenching
II. Structural and Functional Characteristics
A. Comparative Probe Architecture
Parameter cRNA Probes cDNA Probes Oligonucleotide Probes Strandedness Single Double/Single Single Length Range 200-3000 nt 100-5000 nt 12-50 nt Thermal Stability (Tm) 75-90°C 70-85°C 55-75°C Hybridization Efficiency ★★★★★ ★★★☆☆ ★★★★☆ (Fig. 2: Molecular dynamics of probe-target hybridization)
Description: Cryo-EM reconstruction showing cRNA probe (blue) forming A-form helix with target mRNA (gold).B. Signal Transduction Mechanisms
- Direct Labeling:
- Fluorophores (Cy3/Cy5) for real-time imaging
- Biotin-streptavidin amplification systems
- Indirect Detection:
- CRISPR-Cas13 collateral cleavage activation
- Hybridization Chain Reaction (HCR) polymers
III. Source-Specific Production Methodologies
A. cRNA Probe Synthesis
- Critical Components:
- Phage promoter plasmids (pGEM/pBluescript)
- 32P/fluorescent NTP mixes
- Advantages:
- 10-fold higher sensitivity vs. DNA probes
- RNase A digestion compatibility for structure probing
B. Oligonucleotide Probe Engineering
Modification Type Functional Impact Commercial Example 2′-O-methyl Nuclease resistance RNAscope® probes LNA bases Tm increase (2-8°C/nucleotide) Stellaris® FISH Selenophene-uracil X-ray crystallography compatibility Structural biology probes (Fig. 3: Chemically modified nucleotide analogs)
Description: Molecular models comparing standard ribose (gray) with 2′-O-methyl (red) and LNA (blue) configurations.
IV. Application-Optimized Probe Selection
A. Spatial Transcriptomics
- Recommended Probe: Multiplex cRNA probes
- Technology: RNAscope® with pre-amplifier/amplifier system
- Resolution: Single-molecule detection in FFPE tissues
B. Dynamic Live-Cell Imaging
- Recommended Probe: Molecular beacons
- Mechanism:
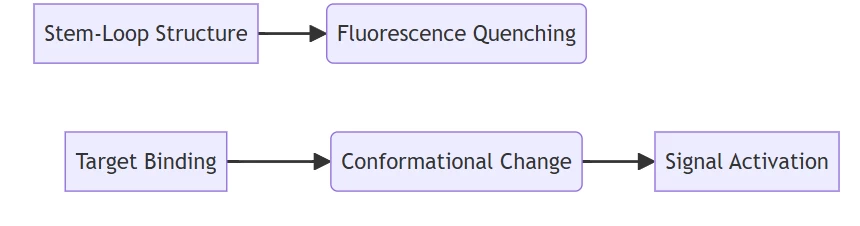
C. Point-of-Care Diagnostics
- Recommended Probe: LNA-modified oligonucleotides
- Platform Integration:
- PrimeFlow™ RNA (Thermo Fisher)
- SARS-CoV-2 detection chips
V. Emerging Hybrid Technologies
A. RNA Origami Nanoprobes
- Structure: Scaffolded 3D assemblies
- Function: Multipathogen capture (e.g., SARS-CoV-2 + influenza)
B. Theranostic Probes
- CRISPR Integration:
- Cas13-cRNA fusions for detection/therapy
- Photocaged Systems:
- 405 nm-activatable probes in neuronal networks
(Fig. 4: CRISPR-RNA theranostic probe)
Description: Schematic showing target RNA cleavage (therapy) and collateral fluorescence activation (diagnostics).
Conclusion: Source-to-Function Paradigm
RNA probe efficacy derives from three interdependent factors:
- Source Fidelity: Phage-derived cRNA > synthetic oligos > cDNA
- Structural Precision: Chemical modifications dictate binding kinetics
- Functional Integration: HCR/CRISPR-enhanced signal amplification
“The evolution from simple hybridization tools to programmable nanodevices represents a quantum leap – contemporary RNA probes now simultaneously map, measure, and modulate cellular RNA landscapes.”
— Nature NanotechnologyOngoing research focuses on in vivo self-assembling probes capable of blood-brain barrier penetration for neurological disorder diagnostics by 2030.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
- (Fig. 1: In vitro transcription schematic)
