 I. Core Process Architecture
I. Core Process Architecture
RNA extraction kits employ systematic biochemical workflows to isolate intact RNA from complex biological matrices while eliminating contaminants. The universal framework comprises four phases:
- Cellular Disruption & Lysis
- Contaminant Removal & RNA Binding
- Matrix Purification & Washing
- Elution & Quality Verification
(Fig. 1: Universal RNA Extraction Workflow)
Description: Circular diagram with color-coded phases: Lysis (red), Binding (blue), Washing (green), Elution (gold). Icons depict tissue homogenization, phase separation, column purification, and spectrophotometry.
II. Phase 1: Lysis & Initial Processing
A. Sample-Specific Disruption Methods
| Sample Type | Lysis Technology | Critical Reagents |
|---|---|---|
| Plant seeds | Mechanical grinding + Buffer S1 (oil/starch separation) | β-mercaptoethanol, chaotropic salts |
| FFPE tissues | Xylene deparaffinization → Proteinase K digestion | Cross-link reversal buffers |
| Viral particles | Immunomagnetic capture + capsid disruption | Viral lysis buffers |
| Exosomes | Immunobead capture → Ethanol precipitation | Anti-exosome antibodies |
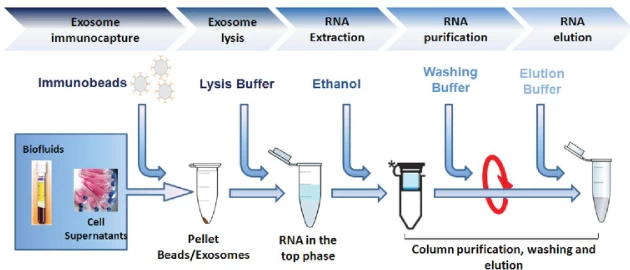
B. Key Innovations
- Simultaneous DNase treatment: gDNA removal during lysis (e.g., DNase I digestion in spin columns)
- Carrier RNA augmentation: MS2 bacteriophage RNA boosts low-yield samples
- Phase-separation chemistry: TRIzol/chloroform isolates RNA in aqueous phase
III. Phase 2: RNA Binding & Contaminant Removal
A. Dominant Binding Technologies
- Silica-Membrane Adsorption (Spin Columns)
- RNA binds to silica at high ionic strength (>4M guanidinium)
- Contaminants flow through during centrifugation
(Fig. 2: Spin-Column Binding Mechanism)
Description: Cross-section showing RNA (blue strands) adhering to silica membrane while proteins/lipids (red/green) pass through.
- Magnetic Bead Capture
- Oligo-dT/silica-coated beads bind RNA
- Magnetic racks isolate RNA-bead complexes
- Direct-zol™ Technology
- Eliminates phenol-chloroform phase separation
- Direct TRIzol lysate application to purification columns
B. Critical Separation Steps
| Contaminant | Removal Method |
|---|---|
| Genomic DNA | On-column DNase digestion |
| Proteins | Ethanol/chaotrope washes |
| Polysaccharides | PVP-40 treatment |
| Lipids | Chloroform extraction |
IV. Phase 3: Rigorous Washing Protocols
A. Universal Wash Sequence
1. **Wash Buffer 1**: High-salt solution removes residual proteins
2. **Wash Buffer 2**: Ethanol-based (70-80%) eliminates salts
3. **Optional DNase Wash**: Column-immobilized DNase digests DNA
Note: Centrifugation at 12,000 rpm ensures complete contaminant removal
B. Specialized Washes
- FFPE samples: Extended 24-hour proteinase digestion
- Plant materials: Double chloroform extraction for starch removal
- Blood samples: Hemoglobin inhibitor cocktails
V. Phase 4: Elution & Quality Control
A. Elution Optimization
- Low-ionic buffers: Nuclease-free water or TE buffer maximizes yield
- Temperature enhancement: 65°C incubation improves RNA solubility
- Volume calibration: 30-50µl balances concentration vs. recovery
B. Quality Verification Metrics
| Parameter | Target Value | Validation Method |
|---|---|---|
| Purity | A260/A280 ≥1.9 | Spectrophotometry |
| Integrity | RIN >7.0 | Bioanalyzer |
| DNA contamination | Ct >35 (no-RT controls) | RT-PCR |
| Yield | >1µg/mg tissue | Fluorometry |
(Fig. 3: QC Electropherogram)
Description: Bioanalyzer trace showing sharp 18S/28S rRNA peaks (RIN=8.2) vs. degraded sample (smear below RIN=5.0).
VI. Technology-Specific Workflows
A. Spin-Column Kits (e.g., GeneJET™)
1. Lyse samples in guanidine-based buffer
2. Load lysate onto silica column → centrifuge (1 min)
3. DNase I treatment on membrane (15 min)
4. Ethanol washes (2× centrifugations)
5. Elute in 30µl nuclease-free water
Processing time: 15 minutes
B. Magnetic Bead Kits (e.g., DP Series)
1. Bind RNA to oligo-dT beads (5 min)
2. Magnet separation → discard supernatant
3. Wash with 80% ethanol (2×)
4. Air-dry beads → elute with water
Throughput: 96 samples in <40 minutes
C. Phase-Separation Kits (e.g., TRIzol-Based)
1. Homogenize in TRIzol → centrifuge (15 min)
2. Transfer aqueous phase to new tube
3. Precipitate with isopropanol → centrifuge
4. Wash pellet with 75% ethanol
5. Air-dry → resuspend in buffer
Key advantage: Unbiased small RNA recovery
VII. Sample-Specific Optimization
| Sample Type | Critical Adaptations | Yield Benchmark |
|---|---|---|
| Plant seeds | Buffer S1 + chloroform extraction | 5µg/100mg |
| Whole blood | Leukocyte stabilization + carrier RNA | 15ng/ml |
| Bacteria | Lysozyme pretreatment + DNase I | 10µg/10^9 cells |
| FFPE | 24h proteinase K → xylene wash | 50% vs. fresh tissue |
VIII. Emerging Innovations
- Single-Column DNA/RNA Separation: Simultaneous isolation from one sample
- Lyophilized Field Kits: Room-temperature stable reagents for point-of-care use
- CRISPR-Assisted Purification: Cas13-RNA complexes for targeted extraction
- Microfluidic Automation: <5-minute processing via integrated chips
“Modern RNA extraction kits transform biological chaos into molecular precision—converting viscous cellular soups into tubes of pure genetic insight.”
— Nature Biotechnology, 2025
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
