 I. Purity Compromises (A260/A280 & A260/A230 Aberrations)
I. Purity Compromises (A260/A280 & A260/A230 Aberrations)
A. Low A260/A280 Ratio (<1.8)
Indicates protein/phenol contamination
- Root Causes:
- Incomplete protein removal during lysis
- Phenol carryover in phase-separation protocols
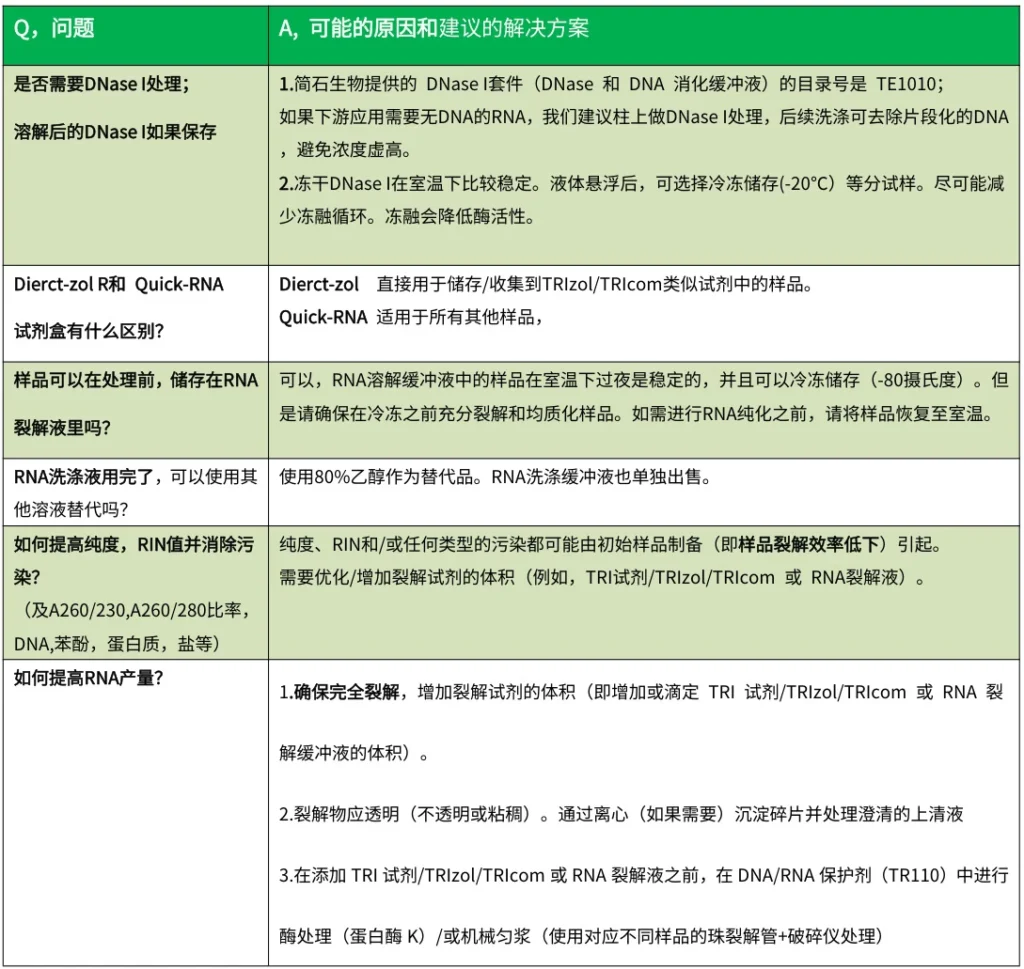
- Insufficient ethanol washes
- Solutions:
- Add 0.1M NaCl → reprecipitate RNA
- Repeat chloroform extraction for phase-separation kits
- Increase Wash Buffer volume by 50%
B. Low A260/A230 Ratio (<1.7)
Suggests carbohydrate/salt contamination
- Root Causes:
- Polysaccharide residuals in plant/fungal samples
- Ethanol precipitation impurities
- Polysaccharide residuals in plant/fungal samples
- Solutions:
- Add 2% PVP-40 to lysis buffer for plants
- Perform extra 75% ethanol wash
- Use nuclease-free water instead of TE buffer for elution
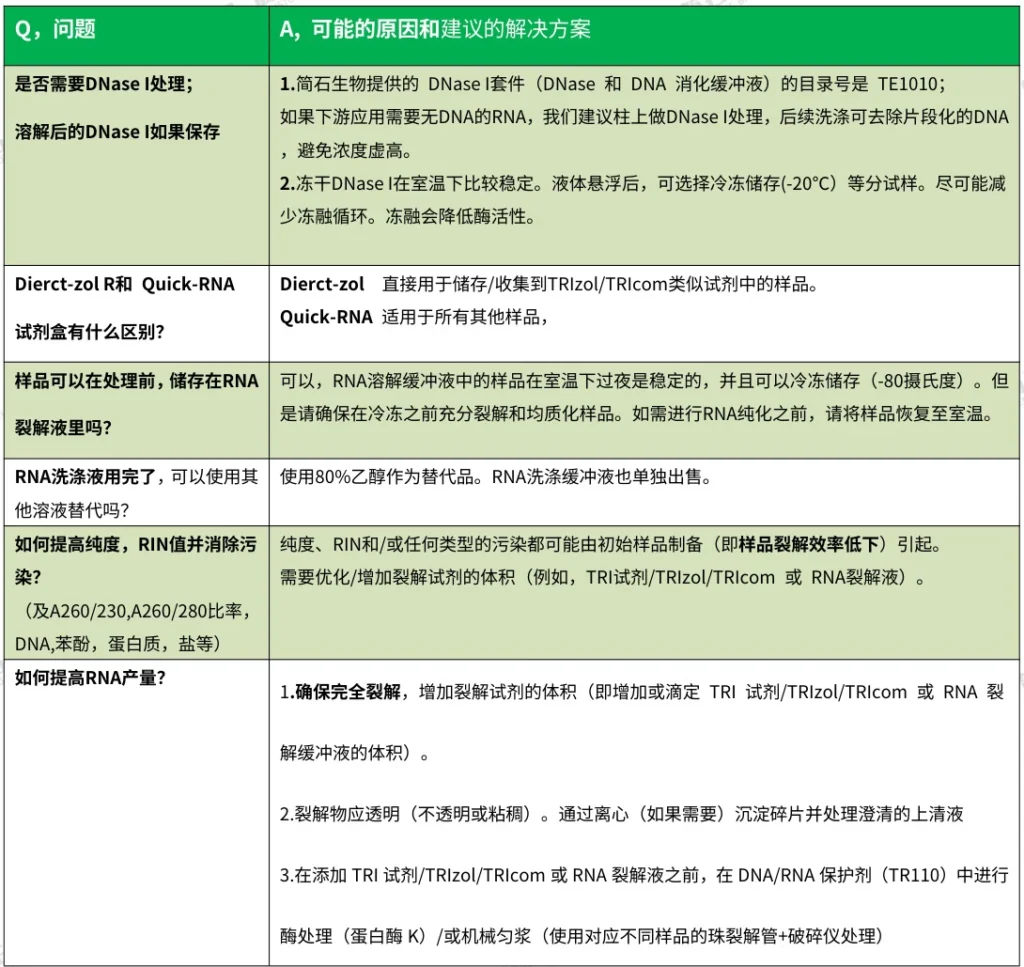
(Fig. 1: Spectrophotometric Purity Diagnostics)
Description: UV trace comparison showing ideal RNA (peaks at 260nm, A260/A280=2.0, A260/A230=2.2) vs. contaminated samples with shifted absorbance ratios.
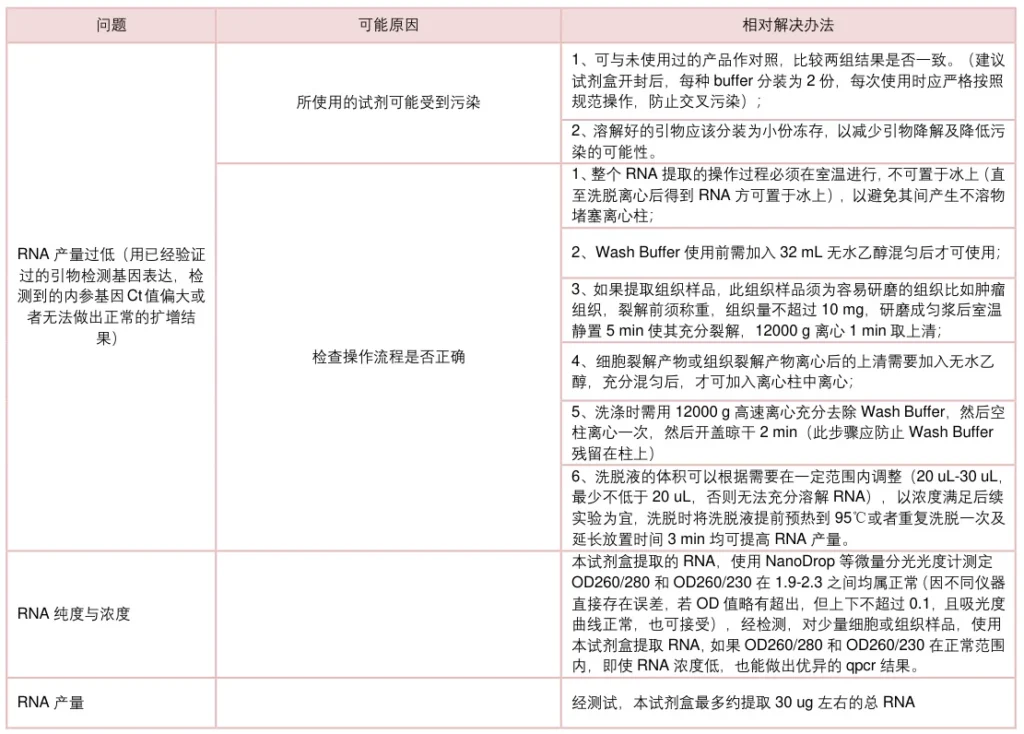
II. Yield Deficiencies
A. Low RNA Recovery
- Root Causes:
- Incomplete cell lysis (bacterial/plant walls)
- RNA adsorption to interphase in phase-separation
- Over-dried silica/magnetic matrices
- Optimization Protocols:
Sample Type Solution Bacteria/Fungi Lysozyme pretreatment (30min/37°C) Plant tissues Liquid N₂ grinding + β-mercaptoethanol FFPE samples 24h proteinase K digestion - Elute with 65°C nuclease-free water
B. Inconsistent Yields
- Root Causes:
- Variable homogenization intensity
- Incorrect ethanol concentration in Wash Buffer
- Solutions:
- Standardize mechanical homogenization (e.g., BeadBeater™)
- Verify ethanol concentration with refractometer
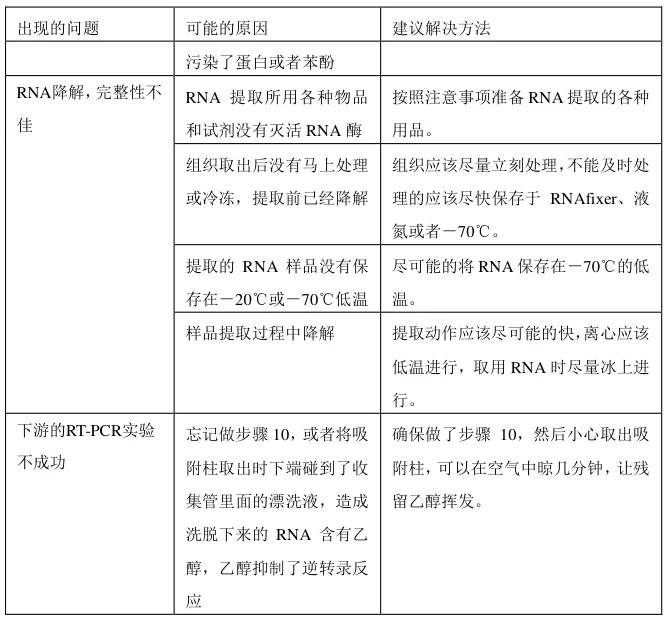
III. RNA Degradation
A. Degradation Indicators
- Bioanalyzer Profile:
- Intact RNA: Sharp 28S/18S rRNA peaks (2:1 ratio, RIN>8.0)
- Degraded RNA: Smear <200nt (RIN<5.0)
(Fig. 2: RNA Integrity Assessment)
Description: Bioanalyzer electropherogram contrasting intact RNA (RIN=8.5) vs. degraded sample (RIN=3.2) with fragmented rRNA.
B. Prevention Workflow
- RNase Inactivation:
- Fresh 0.1% β-mercaptoethanol in lysis buffers
- DEPC-treated consumables + UV workspaces
- Temperature Control:
- Maintain samples at 4°C during processing
- Avoid freeze-thaw cycles (store at -80°C)
IV. Genomic DNA Contamination
A. Detection Methods
- No-RT PCR controls: Ct<35 indicates gDNA contamination
- Gel electrophoresis: High-MW bands
B. Elimination Strategies
| Technology | Protocol |
|---|---|
| Spin-column | On-column DNase I (15min/RT) |
| Magnetic beads | Pre-lysis DNase treatment |
| TRIzol | Acid-phenol re-extraction |
Critical Note:
- DNase I loses 40% activity after 3 freeze-thaw cycles – aliquot single-use portions
V. Sample-Specific Optimization
A. FFPE Tissues
| Challenge | Solution |
|---|---|
| Crosslinking | Xylene deparaffinization → ethanol rehydration |
| Fragmentation | Carrier RNA (e.g., MS2 bacteriophage) |
B. Viral RNA (Plasma/Serum)
- Low Yield:
- Carrier RNA increases recovery by 30-50%
- Tempus™ tubes for transport stabilization
C. Plant/Fungal Samples
- Inhibitor Removal:
- Double chloroform extraction
- 2% PVP-40 in lysis buffer
(Fig. 3: Sample-Specific Workflows)
Description: Illustrated protocols: (A) Plant tissue grinding in liquid N₂, (B) FFPE deparaffinization cascade, (C) Viral RNA capture with magnetic beads.
VI. Operational Best Practices
A. Contamination Prevention
| Risk | Mitigation |
|---|---|
| RNase contamination | Barrier tips + glove changes every 20min |
| Cross-contamination | Dedicated pre-PCR workspace |
B. Critical Parameters
| Step | Requirement | Rationale |
|---|---|---|
| Lysis | Immediate processing | Prevents RNA degradation |
| Ethanol wash | 70-80% concentration | Optimal desalting |
| DNase I | Fresh aliquots | Maintains enzymatic activity |
VII. Emerging Innovations
- CRISPR-Assisted Purification:
- Cas13-RNA complexes for targeted isolation
- Phase-Free Systems:
- Direct-zol™ eliminates phenol-chloroform
- Microfluidic Automation:
- 90-second chip-based extraction
“Mastering RNA extraction transforms biological chaos into molecular precision—where every microliter of eluate contains volumes of unwritten genetic narratives.”
— Journal of Molecular Diagnostics, 2025
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
