 I. Clinical Imperative: PIK3CA as a Therapeutic Biomarker
I. Clinical Imperative: PIK3CA as a Therapeutic Biomarker
PIK3CA mutations—occurring in 20-40% of breast cancers and up to 30% of colorectal/endometrial cancers—constitutively activate the PI3K/AKT/mTOR pathway, driving tumor progression and therapy resistance. These mutations cluster in helical (exon 9: E542K, E545K) and kinase (exon 20: H1047R/L) domains, creating actionable targets for inhibitors like alpelisib. PCR-based detection enables:
- Therapy Selection: Identifying candidates for PI3Kα inhibitors
- Resistance Prediction: Flagging EGFR/HER2 therapy non-responders
- Prognostic Stratification: Correlating mutations with aggressive phenotypes
(Fig. 1: PIK3CA Signaling Pathway)
Description: Gain-of-function mutations (red stars) in PIK3CA exons 7/9/20 drive constitutive PI3K activation, promoting cell survival and proliferation. Alpelisib (blue) selectively inhibits mutant p110α.
II. PCR Methodologies: Balancing Sensitivity and Accessibility
A. Targeted Amplification Technologies
| Platform | Mechanism | Clinical Utility |
|---|---|---|
| ARMS-PCR | Allele-specific primers + hydrolysis probes | Detects 75% of common mutations at ≤1% allele frequency |
| ddPCR | Microfluidic partitioning + TaqMan chemistry | Absolute quantification at 0.1% sensitivity |
| RT-PCR | Real-time fluorescence monitoring | High-throughput screening with <4h turnaround |
B. Detection Coverage Optimization
Therascreen® PIK3CA RGQ PCR Kit (QIAGEN) exemplifies clinical-grade PCR:
- Targeted Exons: 7 (C420R), 9 (E542K/E545A/E545G/E545K/Q546E/Q546R), 20 (H1047L/H1047R)
- Sensitivity: Detects 1 mutant in 100 wild-type alleles
- Sample Compatibility: FFPE, plasma ctDNA, fine-needle aspirates
(Fig. 2: ARMS-PCR Workflow)
Description: DNA extraction → PCR amplification with mutation-specific primers → fluorescence detection. Electropherogram shows E545K mutation peak.
III. Breast Cancer: Evidence-Driven Implementation
A. Metastatic HR+/HER2- Breast Cancer
A 2024 cohort study (n=231) revealed:
- Mutation Prevalence: 39.4% (91/231) harbored PIK3CA mutations
- Dominant Mutations: H1047R (33.3%), E545K (20.9%), E542K (24.2%), H1047L (8.8%)
- Co-mutations: 9.9% had dual PIK3CA mutations
B. Clinical Actionability
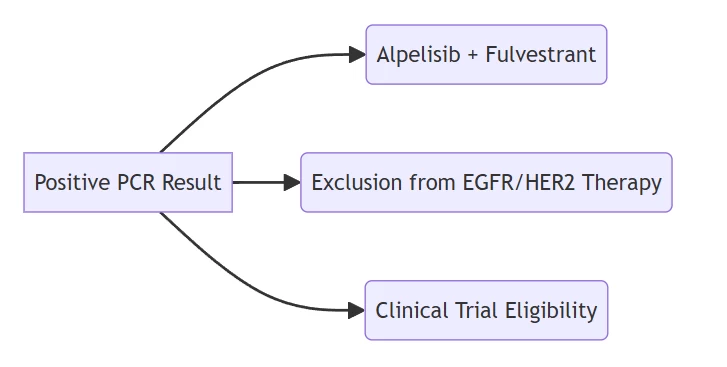
Therapeutic decision tree based on PCR outcomes
IV. Liquid Biopsy Applications: Overcoming Tissue Limitations
A. Plasma ctDNA Analysis
ddPCR Advantages:
- Non-invasive Monitoring: Detects resistance mutations during treatment
- Heterogeneity Capture: Identifies spatially distinct clones
- Dynamic Tracking: Quantifies mutation clearance post-therapy
A 2022 ddPCR assay achieved:
- Multiplex Capacity: 11 mutations in single reaction
- Sensitivity: 2.8-26 mutant copies/reaction
- Clinical Yield: 22.2% positivity in metastatic cases
(Fig. 3: ddPCR Mutation Tracking)
Description: Longitudinal plasma analysis showing H1047R variant allele frequency (VAF) reduction during alpelisib therapy.
V. Global Implementation Frameworks
A. Chinese Expert Consensus (2025)
- Method Selection:
- PCR: First-line for hotspot screening (cost: $50-100/test)
- NGS: Reserved for equivocal PCR results
- Testing Criteria:
Patient Group Testing Recommendation HR+/HER2- metastatic BC Mandatory pre-therapy Early-stage BC Conditional (prognostic assessment) - Sample Priority:
- Primary: FFPE tumor tissue
- Alternative: Plasma ctDNA (55% concordance)
B. Quality Assurance
- Pre-analytical: Tumor cell enrichment >20%
- Analytical: Internal controls for inhibition detection
- Post-analytical: VAF reporting >1% for clinical actionability
VI. Emerging Frontiers and Challenges
A. Multi-Hit Mutation Phenotypes
Prostate cancer data reveals:
- Multi-hit PIK3CA: Associated with elevated TMB/MSI-H status
- Therapeutic Implications: Potential synergy with immunotherapy
B. Technical Innovations
- CRISPR-Enhanced PCR: Improves specificity for rare variants
- AI-Based Interpretation: Predicts mutation functional impact
- Single-Cell PCR: Resolves intratumoral heterogeneity
Conclusion: PCR as the Cornerstone of Precision Oncology
PCR-based PIK3CA profiling demonstrates indispensable value through:
- Clinical Utility: Directing alpelisib therapy in 40% of HR+ breast cancers
- Accessibility: Enabling decentralized testing via standardized kits
- Evolvability: Integrating with liquid biopsy and AI technologies
“Where NGS reveals the genomic landscape, PCR provides the clinical compass—transforming PIK3CA mutations into actionable therapeutic pathways.”
— Journal of Molecular Diagnostics, 2025
Future development prioritizes multi-gene PCR panels (2026) and point-of-care microfluidic systems (2028), with global PCR biomarker testing projected to exceed $5.8B by 2030.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
