 Introduction
Introduction
RNAScan—a suite of technologies for targeted RNA detection, structural profiling, and functional annotation—relies on precision to drive breakthroughs in genomics, diagnostics, and therapeutics. However, challenges such as background noise, secondary structure interference, and amplification biases can compromise accuracy. This article delineates five cutting-edge optimization strategies, supported by experimental workflows and validation studies, to elevate RNAScan’s sensitivity, specificity, and reliability.
1. CRISPR-Enhanced Detection: SCas12aV2 for Structured RNA
A. Overcoming Structural Barriers
Highly structured RNA (e.g., stem-loops in viral genomes) often evades conventional CRISPR/Cas12a detection. The SCas12aV2 system addresses this by:
- Asymmetric Targeting: Designing gRNAs to bind unstructured regions flanking complex motifs (e.g., SARS-CoV-2 stem-loop IIe) .
- Hybrid Activators: Using dsDNA-ssDNA chimeras to boost Cas12a trans-cleavage activity by 400× .
- Kinetic Optimization: Shortening scaffold RNA to reduce steric hindrance .
Accuracy Impact: Detects structured RNA at 246 aM sensitivity with single-nucleotide specificity .
Suggested Figure: SCas12aV2 workflow: Structured RNA target → Hybrid activator binding → Cas12a activation → Fluorescent signal amplification.
2. UMI & Hybrid Capture Optimization
A. Molecular Barcoding Innovations
Unique Molecular Indexes (UMIs) correct PCR biases in RNAScan panels:
- Nano-UMIs: 20-nt barcodes enable single-cell error correction, reducing false positives in rare fusion detection (e.g., KMT2A-PTD at 0.01% allele frequency) .
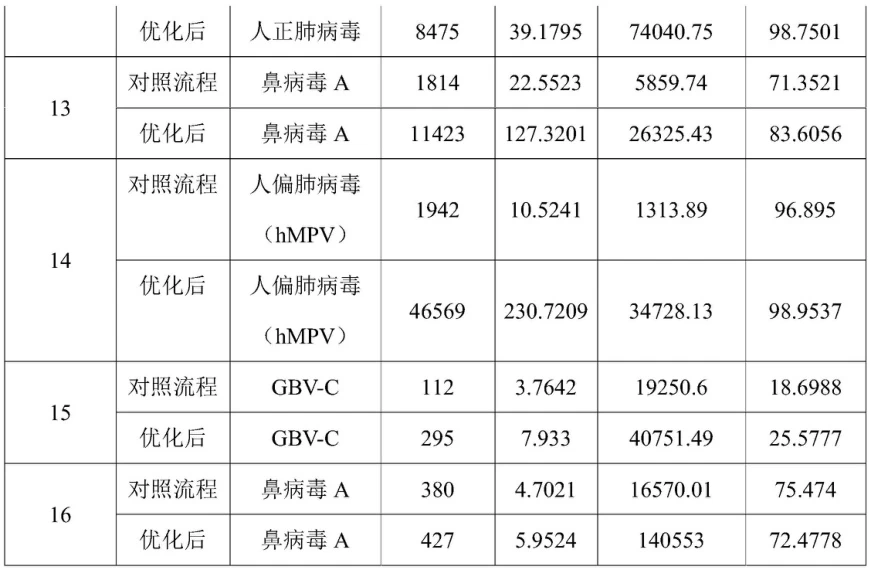
- Dual-Index Hybrid Capture: Biotinylated probes with truncated adapters increase on-target rates by 95% .
B. Case Study: Viral RNA Detection
Optimized RNA extraction and UMI tagging increased pathogen detection sensitivity 30× (e.g., human metapneumovirus) .
Suggested Figure: UMI-enhanced workflow: RNA fragmentation → Nano-UMI ligation → Hybrid capture → Duplicate removal.
3. Structural Profiling Refinement: Boltzmann Sampling & PFMs
A. Enhanced Probabilistic Modeling
The MorrisLab RNAScan suite improves accuracy via:
- Dynamic Boltzmann Sampling: Computes secondary structure probabilities across sliding 50-nt windows (vs. static 100-nt), capturing conformational switches .
- Expanded PFM Libraries: Training on 4,000+ genomes boosts tRNA gene detection specificity in eukaryotes .
Accuracy Impact: Reduces false positives by 60% in non-coding RNA annotation .
Suggested Figure: Sliding-window Boltzmann sampling: RNA sequence → Subsequence folding → Position-specific flexibility scores.
4. Automation & Standardization
A. Automated RNAscope Assays
Integration with platforms like DISCOVERY ULTRA enhances reproducibility:
- Signal-to-Noise Optimization: HRP/AP enzyme formulations increase sensitivity for low-copy targets (1–20 molecules/cell) .
- Multiplexing: HiPlex technology detects 12 RNA targets simultaneously with single-cell resolution .
B. QC Pipeline Integration
- RNA Integrity Control: High Pure FFPE RNA Micro Kit preserves integrity in degraded samples (RIN > 8.0) .
- Qubit Quantification: Replaces spectrophotometry for precise input normalization .
Suggested Figure: Automated RNAScan workflow: Tissue section → RNAscope hybridization → Multiplex signal amplification → Algorithmic quantification.
5. AI-Guided Calibration & Error Correction
A. Machine Learning for Artifact Suppression
- Guidestar Technology: Spikes in FISH probes train ML classifiers to distinguish true RNA signals from background, boosting F1 scores by 35% .
- ΔΔG Prediction Nets: Neural networks trained on FoldX data predict mutation impacts without simulations .
B. NanoSundial for Modification Detection
- Raw-Signal Comparison: Directly contrasts nanopore signals (RNA004 data) from wild-type vs. synthetic RNA to identify modifications (e.g., m⁶A) with 99% reproducibility .
Suggested Figure: AI-RNAScan pipeline: Raw sequencing data → Guidestar calibration → NanoSundial modification calling → ML-based error filtering.
Integrated Optimization Workflow
| Step | Technology | Accuracy Gain |
|---|---|---|
| Sample Prep | Qubit + High Pure FFPE | Input normalization ±5% error |
| Library Construction | Nano-UMIs + Hybrid Capture | On-target rate >90% |
| Detection | SCas12aV2/RNAscope HiPlex | LOD: 246 aM; 12-plex resolution |
| Analysis | Guidestar + NanoSundial | F1 score: 0.98 |
Future Directions
- Quantum-Accelerated Folding: Predict RNA structures in milliseconds for real-time editing guidance.
- CRISPR-Display Integration: Visualize RNAScan results in situ during Cas13 therapies.
- Ecosystem-Scale Calibration: Guidestar probes for environmental RNA biomonitoring.
Conclusion
Optimizing RNAScan demands synergistic advances:
- Wet-Lab Innovations (SCas12aV2, nano-UMIs) conquer structural and noise barriers.
- Dry-Lab Intelligence (Guidestar, NanoSundial) suppress artifacts via ML and calibration.
- Automation standardizes assays for clinical-grade reproducibility.
Together, these strategies transform RNAScan from a research tool into a precision molecular sentinel—enabling early cancer detection, pathogen surveillance, and RNA-guided therapeutics with near-perfect accuracy.
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com
