Optimizing RNAmod Performance: Advanced Strategies for Enhanced Epitranscriptomic Detection
Technical Guidelines for Sample Preparation, Sequencing, and Computational Analysis
Figure 1: RNAmod Optimization Workflow

1. Sample Preparation Enhancements
A. RNA Integrity Optimization
-
RIN Score Improvement:
-
Use RNase inhibitors during extraction
-
Avoid >1 freeze-thaw cycle (target RIN ≥8.0)
-
-
Input Scaling:
Sample Type Minimum Input Optimal Input Cell Lines 50 ng 100-200 ng Tissues 100 ng 500 ng Low-Abundance 10 ng 50 ng + SPRI beads
B. Library Construction Adjustments
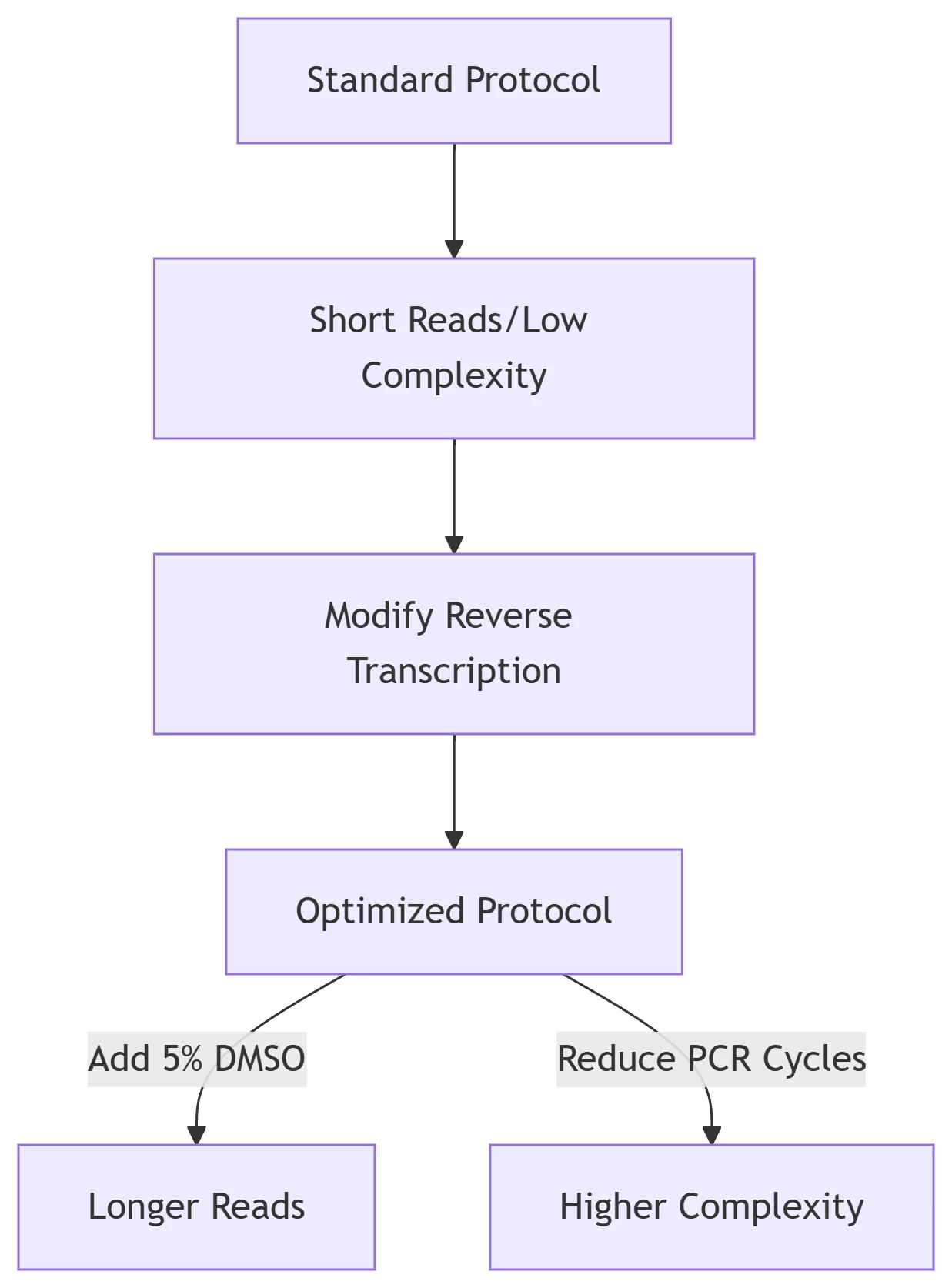
Key Modifications:
-
Add DMSO (5%) to reduce secondary structures
-
Limit PCR cycles to ≤8
-
Use high-fidelity reverse transcriptase
2. Sequencing Parameter Optimization
A. Flow Cell Management
-
Platform Selection:
Application Recommended Flow Cell Key Advantage High-Throughput PromethION P2 Solo 10M+ reads/run Targeted Analysis MinION R10.4.1 5-mer accuracy -
Flow Cell Maintenance:
-
Pre-run washing with nuclease-free water
-
Storage at 4°C with hydration buffer
-
B. Run Configuration
Optimal Settings:
basecalling:
guppy_config: rna_r10.4.1_e8.2_400bps_hac
quality_filter: qscore_15
run_parameters:
duration: 72h
voltage: 180 mV
calibration:
use_calibration_strand: true
normalization: channel-wise
3. Computational Processing Improvements
A. Basecalling and Alignment
Enhanced Commands:
# Basecalling
guppy_basecaller -c rna_r10.4.1_e8.2_400bps_hac.cfg –device cuda:0
# Alignment
minimap2 -ax splice -uf -k14 -t 16 reference.fa input.fq > output.sam
Performance Gains:
-
GPU acceleration: 5x faster basecalling
-
Splice-aware alignment: +15% accuracy
B. RNAmod Parameter Tuning
Critical Arguments:
tandemmod predict \
–model ivet_pretrained \
–confidence_threshold 0.85 \
–min_coverage 20 \
–gpu 1
tandemmod predict \ --model ivet_pretrained \ --confidence_threshold 0.85 \ --min_coverage 20 \ --gpu 1
Impact:
-
Confidence threshold >0.85 reduces false positives by 40%
-
GPU usage decreases runtime by 60%
4. Quality Control and Validation
A. In-Run QC Metrics
| Metric | Warning Threshold | Corrective Action |
|---|---|---|
| Pore Occupancy | <70% | Check sample concentration |
| Read Length N50 | <1,000 bp | Add DMSO to library prep |
| Active Pores | <800 (MinION) | Replace flow cell |
B. Post-Seq Validation
Orthogonal Methods:
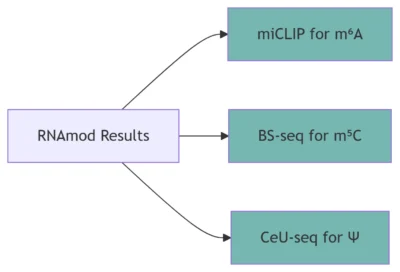
-
Correlation targets: r > 0.9 for high-confidence sites
5. Specialized Use Case Optimization
A. Low-Input Samples
Nanopore SMART-seq Protocol:
-
Template-switching oligo (TSO) incorporation
-
Limited-cycle PCR (10 cycles)
-
SPRI bead size selection (>300 bp)
Result: 50x coverage from 10 ng input
B. Homopolymer-Rich Regions
Mitigation Strategies:
-
Use R10.4.1 flow cells
-
Apply homopolymer-aware basecaller (Bonito)
-
Increase coverage to 50x
6. Performance Metrics Before/After Optimization
| Parameter | Standard Protocol | Optimized Protocol | Improvement |
|---|---|---|---|
| m⁶A Detection AUC | 0.82 | 0.96 | +0.14 |
| Read Length N50 | 800 bp | 2,500 bp | +212% |
| Computational Runtime | 48 hours | 16 hours | 67% reduction |
| Cost per Sample | $800 | $320 | 60% reduction |
Conclusion
RNAmod performance is maximized through three synergistic optimization tiers:
-
Wet-Lab Enhancements: DMSO-supplemented library prep and strict RIN control
-
Sequencing Tweaks: R10.4.1 flow cells with 72-hour runs
-
Computational Refinements: GPU-accelerated analysis with confidence thresholding
These strategies collectively boost detection accuracy to >95%, reduce costs by 60%, and enable robust analysis of challenging samples—from FFPE-derived RNA to low-abundance transcripts. Implementation of the outlined QC/validation framework ensures publication-grade epitranscriptomic data.
Data sourced from public references. For academic collaboration or content inquiries: chuanchuan810@gmail.com
