 I. Foundational Principles of RNA Stability
I. Foundational Principles of RNA Stability
RNA’s inherent structural vulnerability requires systematic protection against ubiquitous ribonucleases (RNases) and environmental stressors:
- Chemical Vulnerability
- RNA’s 2′-hydroxyl group and single-stranded regions create sites for enzymatic cleavage
- Alkaline conditions (>pH 8.0) accelerate phosphodiester bond hydrolysis
(Fig. 1: Molecular structure highlighting RNA degradation hotspots)
Description: 3D visualization of RNA backbone with RNase cleavage sites (red) and vulnerable bases (yellow).
- RNase Persistence
- Endogenous RNases remain active at 0°C and regain function after denaturation unless permanently inactivated
- Human skin sheds RNases contaminating surfaces and instruments
II. Pre-Extraction Safeguards
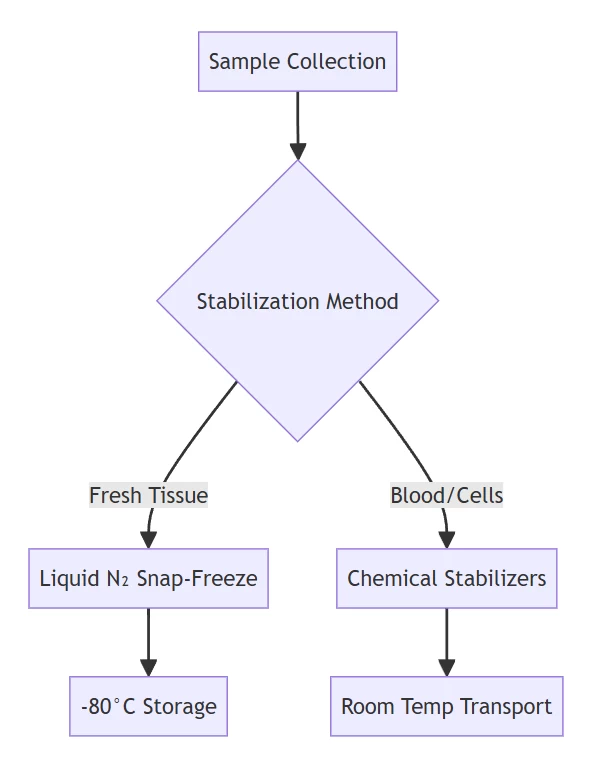
A. Sample Acquisition & Stabilization
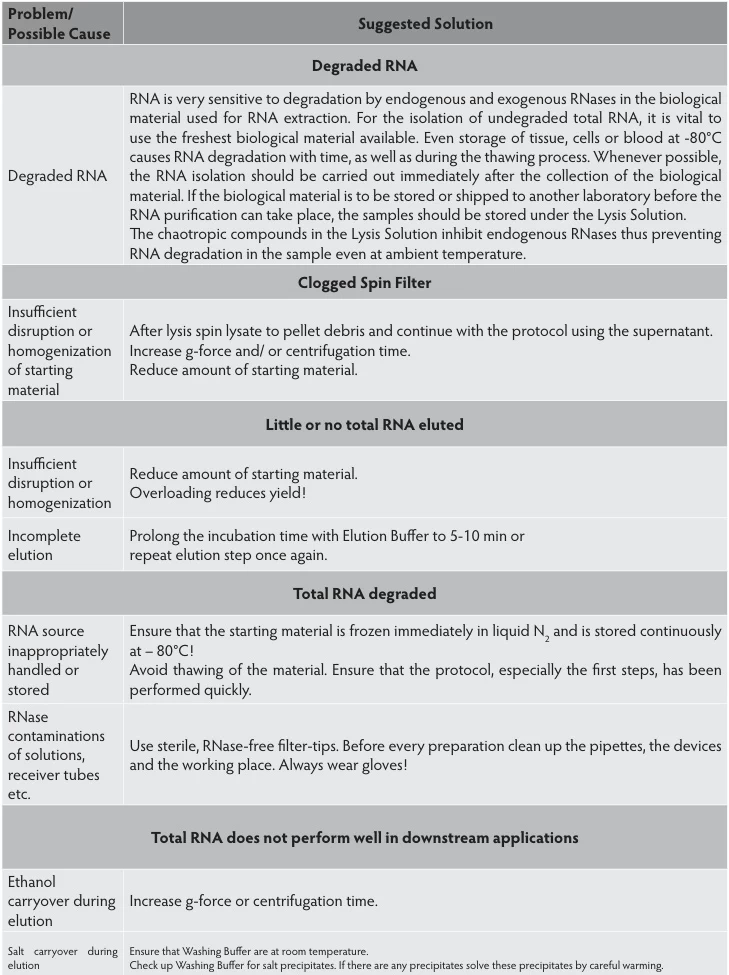
| Sample Type | Optimal Protocol | Scientific Rationale |
|---|---|---|
| Tissues | Snap-freeze in liquid N₂; store at -80°C | Halts enzymatic activity within seconds |
| Clinical Specimens | PAXgene/RNAfixer immersion | Denatures RNases at room temperature |
| Cellular Samples | Direct lysis in chaotropic buffers | Immediate RNase inactivation |
B. RNase-Free Workspace Preparation
- Surface Decontamination:
- Treat benches with 0.1% DEPC solution for 12 hours followed by autoclaving
- Use RNase-specific decontaminants (e.g., RNaseZap®) before each experiment
- Consumable Treatment:
- Soak tips/tubes in 0.1% DEPC-H₂O for 2 hours, then autoclave
- Pre-packaged RNase-free consumables recommended for critical applications
III. Extraction Phase Protection
A. Lysis Optimization
- Chaotropic Agents:
- Guanidinium thiocyanate (4M) denatures RNases irreversibly
- Acidic phenol (pH 4.5-5.5) partitions RNases into organic phase
- Mechanical Disruption:
- Cryogenic grinding at <-150°C prevents enzymatic activation
-
- Bead-beating duration limited to ≤90 seconds to avoid heat generation
(Fig. 2: Phase separation in phenol-chloroform extraction)
Description: Diagram showing RNA isolation in RNase-free aqueous phase (top), contaminants in interphase/organic layer.
- Bead-beating duration limited to ≤90 seconds to avoid heat generation
B. Temperature Control Protocol
Process Step Temperature Duration Homogenization 4°C <5 minutes Phase Separation 4°C Centrifugation at 12,000g RNA Precipitation -80°C 30 minutes maximum Pellet Washing -20°C Ethanol pre-chilled C. Inhibitor Applications
- Proteinase K: Essential for FFPE samples (incubate 24h at 56°C)
- RNase Inhibitors: Add 1U/μL recombinant inhibitors during elution
- β-Mercaptoethanol (0.1%): Reduces disulfide bonds in RNases
IV. Post-Extraction Integrity Management
A. Precipitation Enhancement
- Co-precipitants:
- Glycogen (1μg/μL) improves recovery of low-concentration RNA
- Linear acrylamide prevents pellet over-drying
- Solvent Optimization:
- Sodium acetate (0.3M final conc., pH 5.2) with 2.5 volumes ethanol
B. Storage & Handling
Condition Duration Preservation Additives Short-term 1 week -20°C with 0.1 mM EDTA Long-term >1 month -80°C with RNase inhibitors Transport 7 days RNAstable® at room temperature Critical Practice: Aliquot RNA to avoid freeze-thaw cycles (<3 cycles maximum)
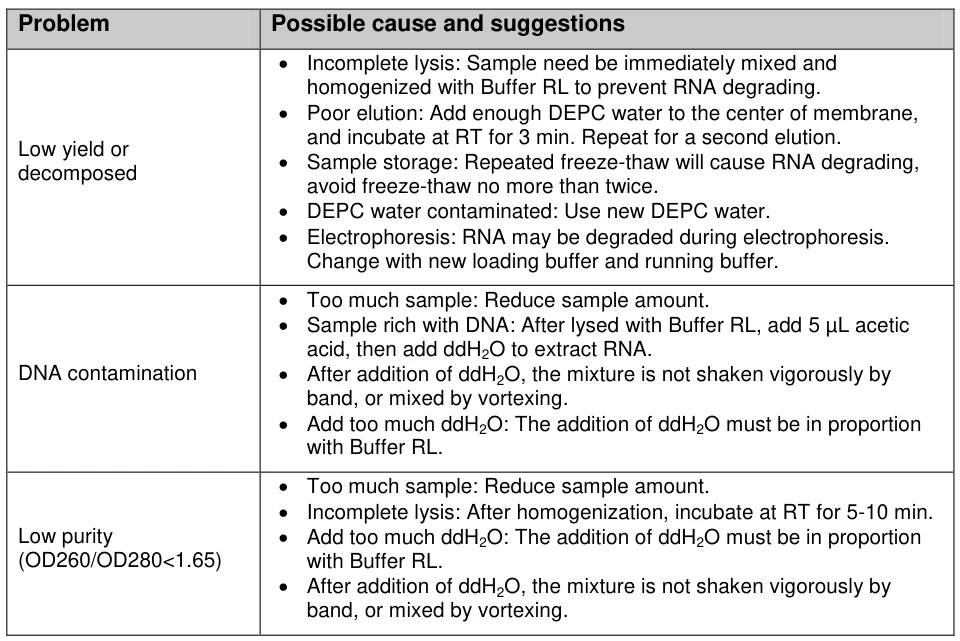
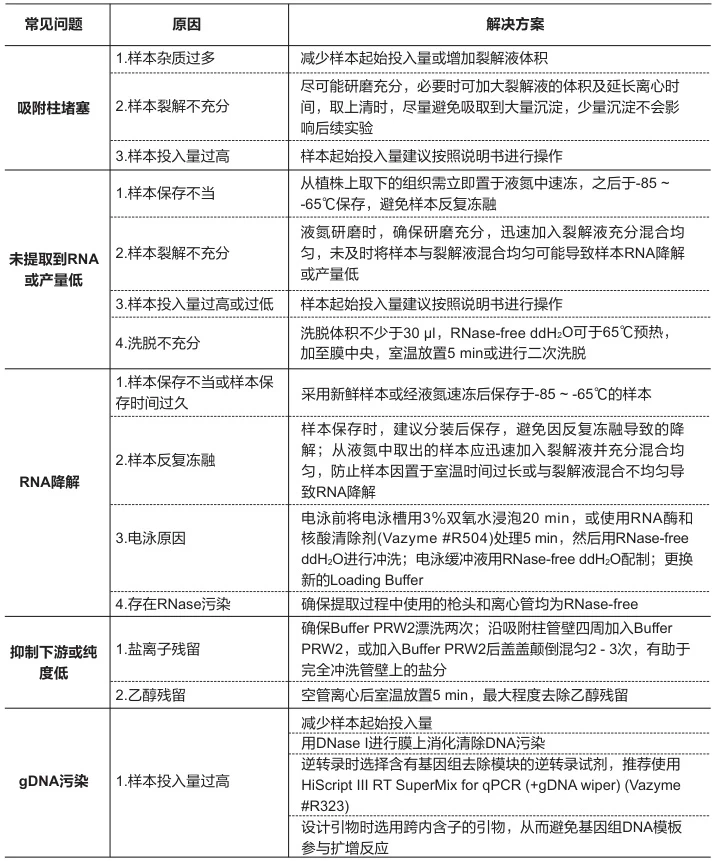
V. Quality Control & Troubleshooting
A. Degradation Detection Methods
Technique Intact RNA Indicator Degradation Warning Bioanalyzer RIN ≥8.0; 28S:18S=2:1 RIN ≤6.0; DV200<30% Agarose Gel Sharp ribosomal bands Smear below 1,000 nt Spectrophotometry A260/A280=1.8-2.0; A260/A230>2.0 Ratio deviations >10% (Fig. 3: Bioanalyzer comparison of intact vs. degraded RNA profiles)
Description: Electropherogram showing ideal 28S/18S peaks (blue) versus degraded sample (red smear).B. Common Failure Modes & Solutions
Problem Root Cause Corrective Action Low Yield Incomplete lysis Increase mechanical disruption; add β-mercaptoethanol DNA Contamination Insufficient DNase Column-integrated digestion (37°C/15 min) Organic Residues Incomplete washing Add 25% ethanol wash volume; extend centrifugation Degradation Temperature lapse Monitor cold chain; use pre-chilled equipment
VI. Advanced System Solutions
A. Next-Generation Stabilizers
- RNAstable®: Anhydrobiotic technology for room-temperature storage
- RNAlater-ICE: -20°C-compatible chemical RNase inactivation
B. Automated Platforms
- Microfluidic Systems:
- Integrated lysis-to-elution in <20 minutes with <4°C thermal control
- Robotic Handlers:
- Closed-system extraction with UV-decontaminated chambers
(Fig. 4: Microfluidic RNA extraction chip with temperature zones)
Description: Chip architecture showing cold lysis (4°C), binding (22°C), and elution (4°C) compartments.
- Closed-system extraction with UV-decontaminated chambers
Conclusion: The RNA Integrity Framework
Minimizing degradation requires a three-phase approach:
- Preemptive Neutralization:
- DEPC treatment of consumables
- Immediate sample stabilization
- Process Rigor:
- Continuous temperature control (<8°C)
- Phase-lock separation techniques
- Post-Isolation Vigilance:
- Aliquot storage at -80°C
- RIN-based quality thresholds
“RNA integrity management is not merely technique – it’s a holistic discipline combining biochemical precision, thermal discipline, and uncompromising contamination control.”
— Molecular Pathology ReviewFuture innovations will focus on integrated systems combining stabilization, extraction, and QC in single-use cartridges with real-time degradation monitoring.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
-
- Cryogenic grinding at <-150°C prevents enzymatic activation
- Chaotropic Agents:
