 I. Foundational Principles for RNA Integrity Preservation
I. Foundational Principles for RNA Integrity Preservation
Successful RNA extraction hinges on preventing degradation while maximizing yield through rigorous RNase control and sample stabilization:
- RNase Elimination Protocols
- Treat all surfaces with RNase decontamination sprays (e.g., RNaseZap®) and use certified RNase-free consumables
- Employ chaotropic agents (guanidinium thiocyanate or phenol-based solutions) during lysis to irreversibly denature RNases
(Fig. 1: Guanidinium ions disrupting RNase tertiary structure)
Description: Molecular visualization showing chaotropic agents denaturing RNase active sites.
- Sample Stabilization Techniques
- Chemical stabilizers preserve RNA integrity for 1 week at room temperature
II. Sample-Specific Optimization Strategies
A. Specialized Lysis Protocols
Sample Type Optimal Approach Critical Modifications Plant Tissues CTAB buffer + 2% β-mercaptoethanol Removes polysaccharides/polyphenols FFPE Samples Extended Proteinase K digestion (24h at 56°C) Reverses formaldehyde crosslinks Adipose/Lipid-Rich Double-volume chloroform washes Eliminates lipid interference Single-Cell/Microbial Mechanical disruption + enzymatic lysis Lysozyme pretreatment for Gram+ bacteria 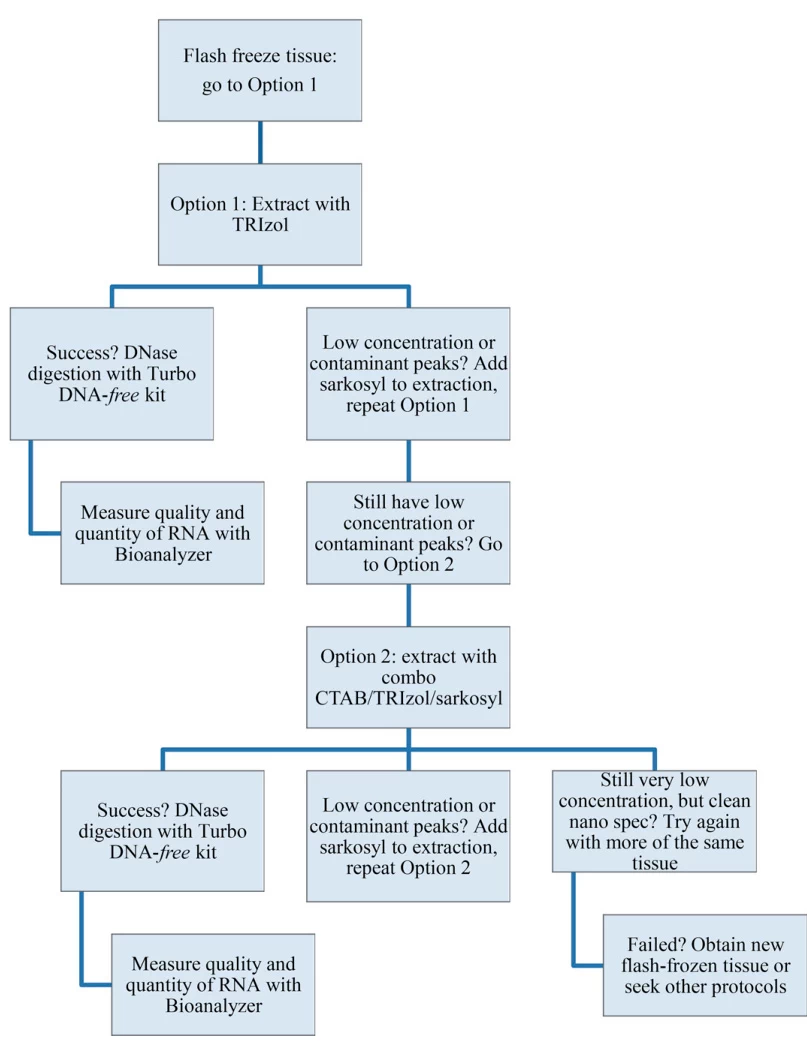
B. Homogenization Enhancement
- Cryogenic Grinding: Pre-cool mortars/pestles in liquid N₂ for brittle fracture homogenization
- Bead-Based Disruption: Use 0.5mm zirconia beads with high-frequency vibration (≥30 Hz) for 90 seconds
III. Extraction Methodology Selection
A. Comparative Matrix of Core Techniques
Method Yield Processing Time Best Applications TRIzol®/Chloroform High 60-90 min Total RNA from diverse samples Silica Columns Medium-High 30 min Clinical diagnostics; DNase-treated RNA Magnetic Beads Medium 20 min High-throughput automation Direct-zol™ High 7 min NGS-ready RNA; microRNAs (Fig. 2: Phase separation dynamics in phenol-chloroform extraction)
Description: Layered solution showing RNA in aqueous phase (top), DNA/protein at interphase, and organic phase (bottom).B. Modified Protocols for Challenging Samples
- Polysaccharide-Rich Plants:
- Add 2% PVP-40 to CTAB buffer during homogenization
- Implement sarkosyl-enhanced TRIzol® extraction for recalcitrant species
- Low-Input Samples:
- Use 1 µg glycogen or linear acrylamide as co-precipitant
- Reduce elution volume to ≤30 µL RNase-free water
IV. Contaminant Removal & Purification
A. DNA Elimination Techniques
- Column-Integrated DNase: On-membrane digestion during silica purification (37°C, 15 min)
- Solution-Phase Treatment: Turbo DNase incubation followed by acid-phenol extraction
B. Organic Contaminant Control
A260/A230 Ratio Interpretation Corrective Action <1.8 Polysaccharide/phenol contamination Increase ethanol wash volume by 25% >2.0 Optimal purity Proceed to downstream applications
V. Quality Control & Integrity Assessment
A. Spectrophotometric Standards
Parameter Optimal Value Significance A260/A280 1.8–2.0 Protein-free RNA A260/A230 >2.0 Absence of organics B. Advanced Integrity Analysis
- Bioanalyzer Profiling:
- Require RIN >7 for RNA-seq applications
- DV200 >30% for FFPE samples (superior to RIN)
(Fig. 3: Bioanalyzer trace comparing intact vs. degraded RNA)
Description: Electropherogram showing distinct 28S/18S peaks (RIN=9.1) versus degradation smear.
VI. Storage & Stability Optimization
A. Temperature Management
Duration Storage Condition Preservation Additives Short-term (1 week) -20°C 0.1 mM EDTA Long-term (>1 month) -80°C RNase inhibitors (e.g., RNAsin®) B. Freeze-Thaw Mitigation
- Aliquot RNA in single-use volumes (≥10 µL)
- Use cryoprotectants like trehalose for −80°C storage
VII. Emerging Innovations
- Microfluidic Platforms:
- Integrated lysis/purification in picoliter chambers (10-min workflow)
- Automated High-Throughput Systems:
- Robotic liquid handlers with temperature-controlled centrifugation
- Spatial Transcriptomics Integration:
- Barcoded oligo-dT arrays for in situ RNA capture
(Fig. 4: Microfluidic chip architecture for automated RNA extraction)
Description: Microchannel network showing cell lysis, binding, and elution zones.
Conclusion: The RNA Extraction Optimization Framework
Maximize yield and integrity through:
- Pre-extraction Stabilization: Snap-freezing or chemical preservation
- Sample-Tailored Lysis: CTAB for plants, extended digestion for FFPE
- Method Selection: TRIzol® for versatility, Direct-zol™ for speed
- Contaminant-Specific Purification: DNase treatment, enhanced ethanol washes
- Integrity-Centric Storage: Aliquotting at −80°C with cryoprotectants
“RNA extraction remains the critical gateway to molecular insights – where meticulous optimization separates actionable data from analytical artifacts.”
— Genomic Technology ReviewFuture advancements will focus on integrated systems combining extraction, quantification, and library prep in closed microfluidic platforms.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
