Mechanisms of Transcription Factor-Mediated RNA Polymerase Recruitment to Promoters
A Structural and Functional Analysis

1. Introduction: The Central Role of Transcription Factors (TFs)
Transcription factors are sequence-specific DNA-binding proteins that recognize cis-regulatory elements in gene promoters, enabling RNA polymerase (Pol) II—incapable of independently binding or unwinding DNA—to locate and initiate transcription14. In eukaryotes, this process requires six general transcription factors (GTFs: TFIID, TFIIA, TFIIB, TFIIF, TFIIE, TFIIH) to form the pre-initiation complex (PIC)18.
2. TF-Promoter Recognition: Sequence-Specific Binding
Key DNA Motifs
-
Core Promoter Elements:
-
TATA box (consensus: TATAWAW) recognized by TBP (TATA-binding protein subunit of TFIID)26.
-
Initiator (Inr) and downstream promoter element (DPE) bound by TAF subunits of TFIID6.
-
-
Proximal Elements:
-
GC/CAAT boxes bound by SP1, NF-Y, etc.6.
-
Structural Basis of Recognition
Transcription factors employ DNA-binding domains (DBDs) to engage specific sequences:
-
Zinc fingers (e.g., SP1)
-
Helix-turn-helix (e.g., homeodomain TFs)
-
Basic leucine zippers (e.g., CREB)
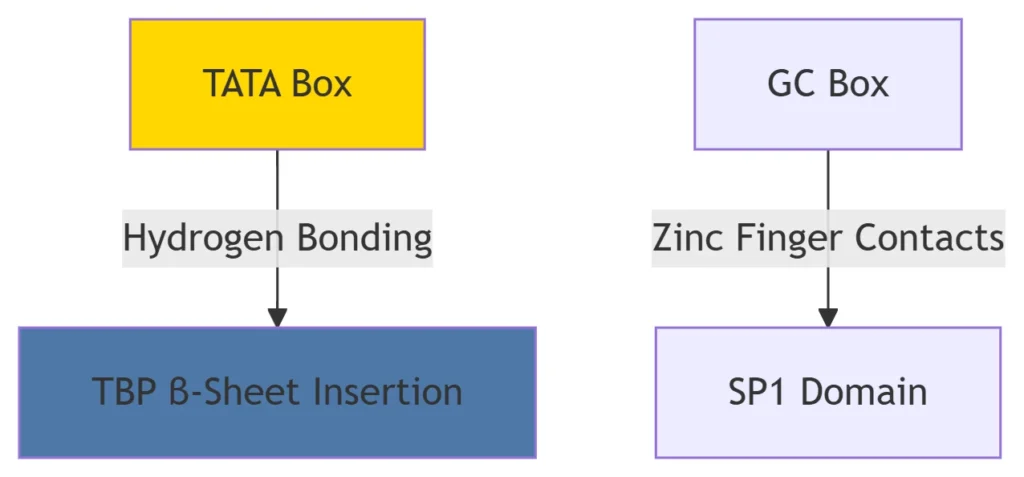
TF-DNA interactions involve shape complementarity and chemical bonding, enabling nanomolar-affinity binding6.
3. PIC Assembly: Stepwise Recruitment of Pol II
Assembly Sequence
-
TFIID Binding:
-
TBP bends DNA by 80°, creating a scaffold for other GTFs18.
-
-
TFIIA/B Recruitment:
-
TFIIB bridges TFIID and Pol II, positioning its B-reader helix at the transcription start site (TSS)48.
-
-
Pol II-TFIIF Entry:
-
TFIIF stabilizes Pol II binding and prevents non-specific DNA interactions1.
-
-
TFIIE/H Incorporation:
-
TFIIH uses ATP-dependent helicase activity to unwind DNA, forming a 14-bp transcription bubble14.
-
Structural Transitions

4. TFIIH-Mediated DNA Melting and Pol II Activation
Energy-Dependent DNA Unwinding:
-
TFIIH hydrolyzes ATP to translocate downstream DNA, generating negative supercoiling that destabilizes duplex DNA18.
Transcription Bubble Formation:
-
Unwound template strand enters Pol II’s active-site cleft, positioning the TSS for initiation4.
5. Transition to Elongation: Promoter Escape
Key Structural Changes
-
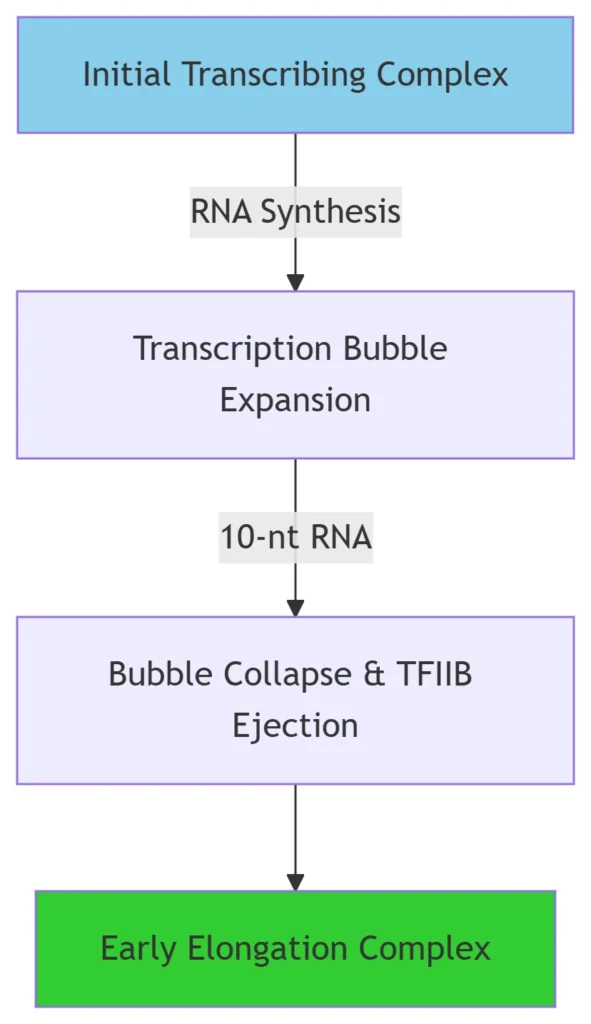
Abortive Initiation:
-
Pol II synthesizes short (2–9-nt) RNAs while anchored to GTFs18.
-
-
Promoter Clearance:
-
At 10-nt RNA synthesis, NTP hydrolysis drives RNA-DNA hybrid translocation, rupturing the TFIIB-Pol II interface and collapsing the transcription bubble from 21 nt to 11 nt48.
-
-
GTF Dissociation:
-
TFIIB detachment allows Pol II to enter processive elongation14.
-
Structural Visualization
6. Polymerase-Specific TF Systems
RNA Polymerase Transcription Factors Target Promoters Pol II GTFs (TFIID, TFIIH, etc.) Protein-coding genes Pol III SNAPc, TFIIIB U6 snRNA, tRNA, 5S rRNA Pol I UBF, SL1 Ribosomal RNA genes Pol III Example:
-
SNAPc complex binds the proximal sequence element (PSE) in U6 snRNA promoters, recruiting TFIIIB and Pol III to form a closed PIC that spontaneously opens DNA5.
7. Functional Implications
Regulatory Precision
-
TFs enable tissue-specific gene expression by responding to signaling pathways (e.g., NF-κB in inflammation)67.
-
Mutations in TF-DNA interfaces cause diseases (e.g., TBP defects in spinocerebellar ataxia)1.
Therapeutic Targeting
-
Small molecules disrupting oncogenic TF-DNA interactions (e.g., STAT3 inhibitors) are in clinical trials6.
Conclusion
Transcription factors serve as molecular matchmakers that recognize promoter signatures, recruit Pol II via PIC assembly, and orchestrate DNA unwinding and promoter escape. Their sequence-specific binding ensures precise transcription initiation, while dynamic PIC remodeling enables the transition to elongation. Recent structural insights—particularly the visualization of TFIIB ejection during promoter escape—highlight how TFs resolve the conflict between stable initiation and efficient elongation18. Deciphering these mechanisms advances gene therapy and drug development for transcription-linked diseases.
Data sourced from public references including:
-
Xu et al. Science (2023) on PIC dynamics148
-
Eukaryotic transcription biochemistry reviews26
-
Structural databases (RCSB PDB: 7RZS, 8H2U)
For academic collaboration or content inquiries: chuanchuan810@gmail.com
-
