Mechanisms of RNA Polymerase Promoter Localization: Molecular Targeting Strategies
A Structural and Functional Analysis
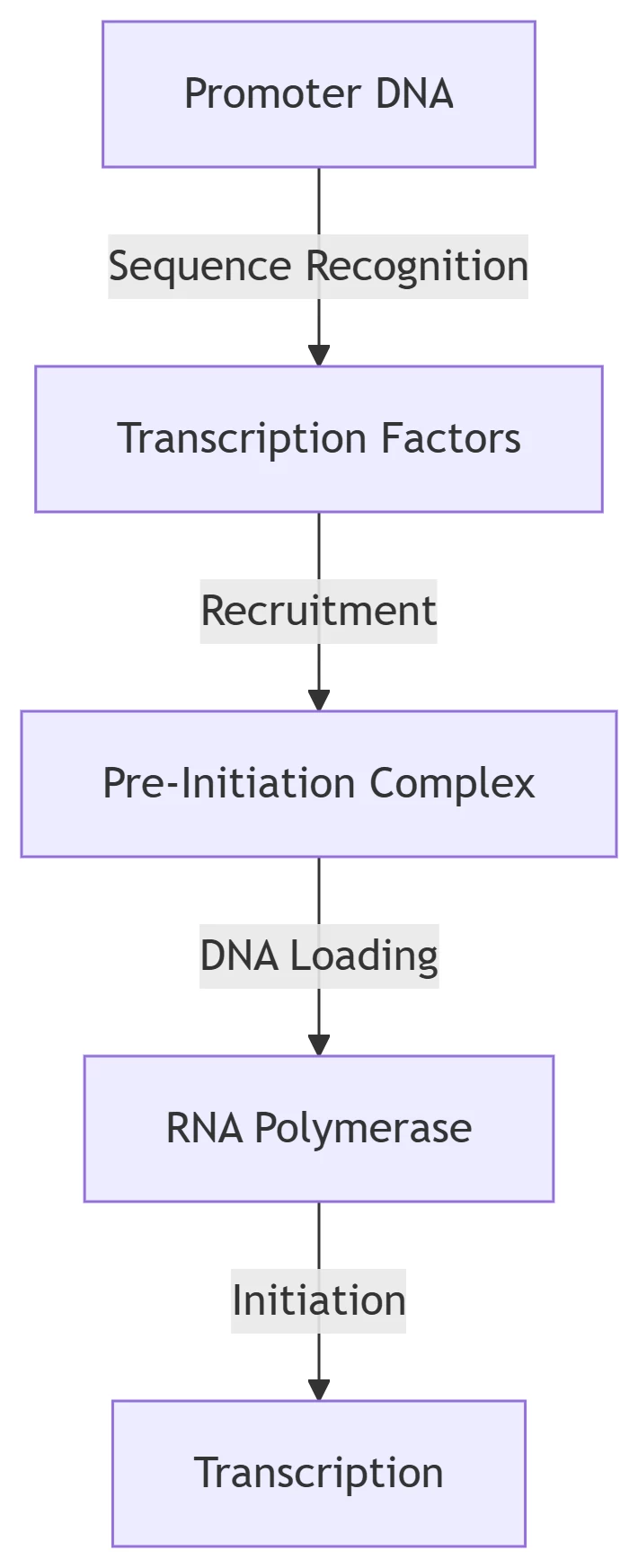
1. Introduction: The Promoter Targeting Problem
RNA polymerases (RNAPs) cannot independently locate promoters among vast genomic DNA. In eukaryotes, RNAP II requires 6 general transcription factors (GTFs) for promoter localization, while bacterial RNAP utilizes σ factors. Key promoter elements include:
-
TATA box (eukaryotes: TATAAA)
-
-10 element (bacteria: TATAAT)
-
Initiator (Inr) sequence
-
Downstream Promoter Element (DPE)
2. Stepwise Localization Mechanisms
A. Eukaryotic System (RNAP II)
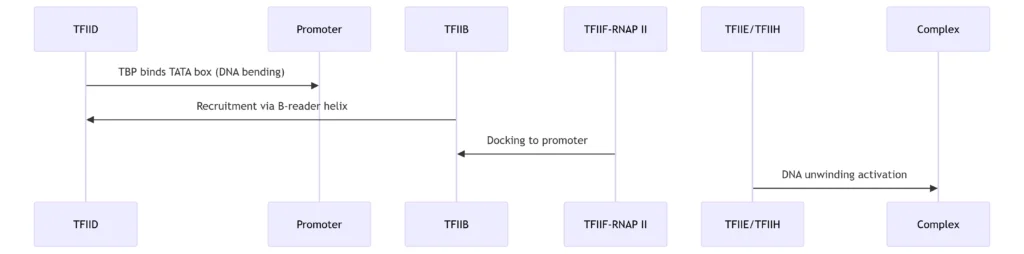
Key Steps:
-
TFIID Recognition: TATA-binding protein (TBP) distorts DNA (80° bend)
-
TFIIB Anchoring: Positions RNAP II at transcription start site (TSS)
-
TFIIF Chaperoning: Escorts RNAP II to PIC while preventing non-specific binding
B. Bacterial System (σ Factor-Dependent)
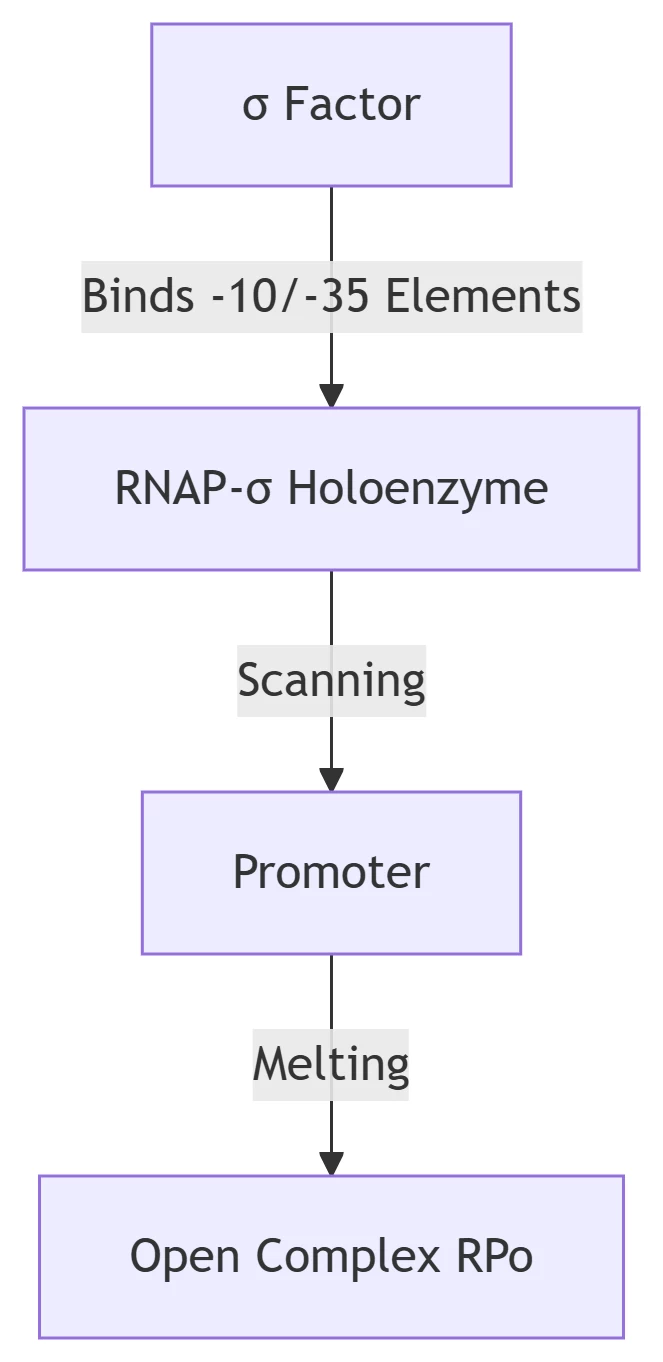
-
σ<sup>70</sup> Domain Structure:
-
Region 2.4: Recognizes -10 element (TATAAT)
-
Region 4.2: Binds -35 element (TTGACA)
-
Linker region: Facilitates DNA melting
-
3. Structural Basis of Recognition
DNA-Protein Interfaces
| Component | Target Sequence | Binding Domain | Affinity |
|---|---|---|---|
| TBP (Eukaryotes) | TATA box | β-sheet saddle | K<sub>d</sub> = 1-10 nM |
| σ<sup>70</sup> (Bacteria) | -10 element | Helix-turn-helix | K<sub>d</sub> = 0.1-1 μM |
| TFIIB (Eukaryotes) | BRE element | Zinc ribbon/B-core | K<sub>d</sub> = 10-100 nM |
DNA Bending Mechanics

TBP inserts phenylalanine residues into minor groove, unwinding DNA and creating protein-docking surfaces.
4. Chromatin Accessibility Strategies
Nucleosome Remodeling
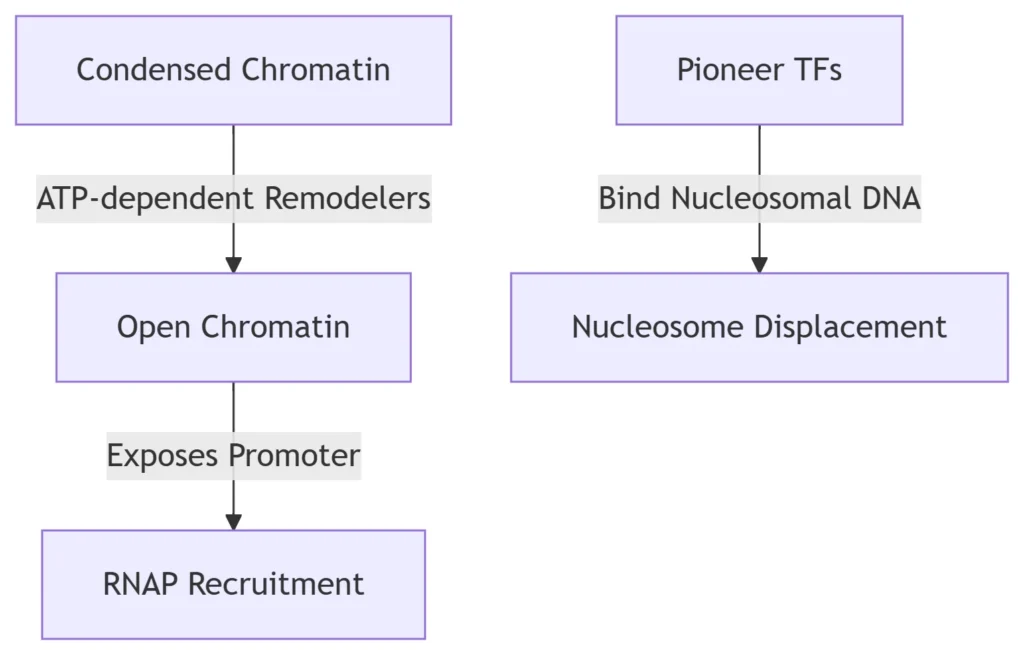
Key Regulators:
-
SWI/SNF complexes: Slide nucleosomes away from promoters
-
Histone acetyltransferases (HATs): Neutralize positive charges
5. Directed Search Mechanisms
Facilitated Diffusion
RNAP locates promoters through:
-
1D Sliding: Linear diffusion along DNA (speed: 500 bp/ms)
-
Hopping: Microscopic dissociation-reassociation
-
Intersegment Transfer: Jumping between DNA segments
Search Optimization:
-
Reduced dimensionality search increases efficiency 100-fold vs 3D diffusion
Bacterial RNAP Search Parameters
| Parameter | Value |
|---|---|
| Diffusion coefficient | 0.04 μm²/s |
| Sliding distance | 45-60 bp |
| Target acquisition | <5 seconds/gene |
6. Transition to Initiation
Open Complex Formation
Energy Requirements:
-
ATP hydrolysis by TFIIH (eukaryotes)
-
Spontaneous in AT-rich bacterial promoters
7. Regulatory Variations Across Domains
| System | Localization Factor | Promoter Architecture | Melting Mechanism |
|---|---|---|---|
| Eukaryotes | GTFs (TFIID/TFIIB) | Modular (TATA/Inr/DPE) | TFIIH ATPase-dependent |
| Bacteria | σ factor | -10/-35 elements | Spontaneous |
| Archaea | TBP/TFB | TATA box/BRE | Hybrid mechanism |
Conclusion
RNA polymerase locates promoters through multi-stage targeting:
-
Sequence Recognition: TBP/σ factors identify core promoter elements
-
Chromatin Navigation: Pioneer TFs and remodelers expose regulatory regions
-
Directed Diffusion: 1D sliding accelerates search kinetics
-
Complex Assembly: GTFs position RNAP at transcription start sites
This hierarchical targeting achieves remarkable precision—eukaryotic RNAP II initiates transcription within ±1 bp of designated start sites despite genomic complexity. Bacterial RNAP-σ holoenzyme locates promoters within seconds through optimized search strategies. Structural insights from cryo-EM (e.g., human PIC at 2.8 Å resolution) continue to reveal dynamic assembly mechanisms with implications for gene therapy and antimicrobial drug design.
Data sourced from public references including:
-
Cramer P. Structural Biology of RNA Polymerase II (Annu Rev Biochem, 2019)
-
Saecker R.M. et al. Mechanism of Bacterial Transcription Initiation (Science, 2021)
-
RCSB PDB structures: 6TWR (human PIC), 6ALF (bacterial RNAP)
-
ENCODE Consortium Promoter Atlas
For academic collaboration or content inquiries: chuanchuan810@gmail.com
