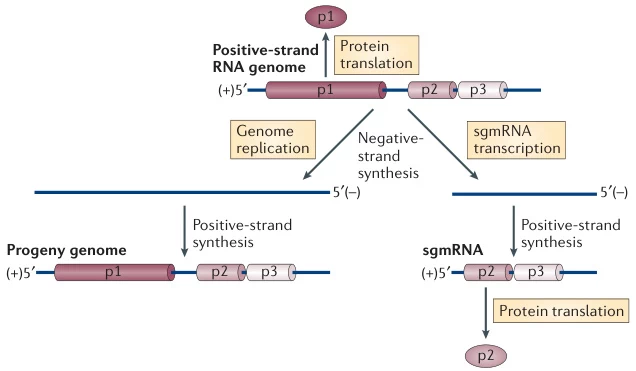
 I. Molecular Framework of Replication Initiation
I. Molecular Framework of Replication Initiation
Positive-sense RNA (+ssRNA) genomes serve dual roles upon host cell entry:
- Immediate Translation Template: Direct synthesis of viral replicase complexes (RdRp, helicases, cofactors)
- Replication Blueprint: Formation of membrane-bound replication organelles (ROs) where negative-strand RNA synthesis occurs
(Fig. 1: Replication Organelle Assembly)
Description: Endoplasmic reticulum membranes (yellow) invaginating to form double-membrane vesicles (DMVs). Viral +ssRNA (blue) recruits host proteins (purple) and RdRp (red) to initiate replication.
II. Stepwise Mechanism of Negative-Strand Synthesis
Step 1: Replicase Complex Assembly
- Structural Requirements:
- cis-acting RNA elements: Conserved stem-loops in 5’/3′ UTRs (e.g., coronavirus TRS-L) serve as RdRp loading platforms
- Host factors: hnRNPs stabilize RNA structures for replicase binding
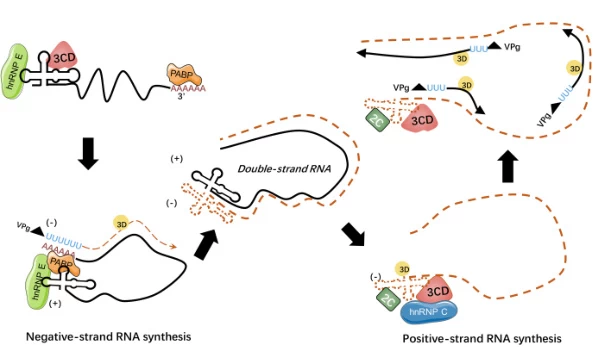
Step 2: Template Activation
- Genome Uncoating: Viral helicases unwind secondary structures in +ssRNA
- Replication Fork Formation: RdRp binds template at precise initiation sites (e.g., PepMV hp2 stem-loop)
Step 3: Negative-Strand Elongation
- Processive Polymerization: RdRp synthesizes complementary RNA in 5’→3′ direction
- Template Switching: Discontinuous transcription at TRS-B sites generates subgenomic RNAs
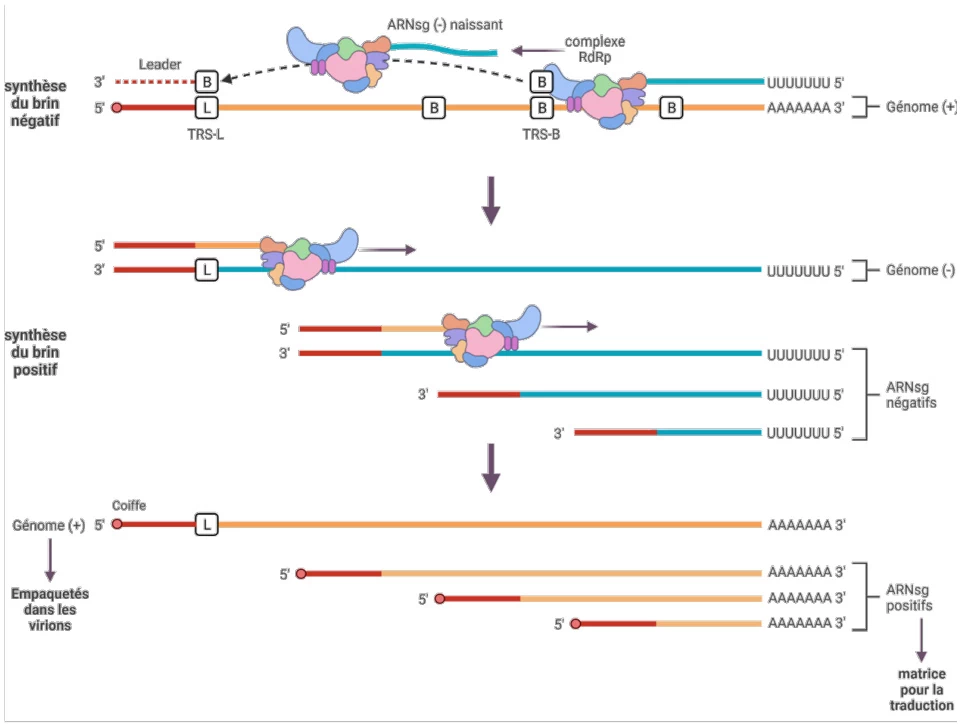
(Fig. 2: Negative-Strand Synthesis Pathway)
Description: RdRp (red) bound to +ssRNA template (blue) synthesizing complementary negative-strand RNA (green). Insets show conserved stem-loop structures directing initiation.
III. Key Molecular Players
| Component | Function | Viral Example |
|---|---|---|
| RdRp Complex | Catalytic RNA synthesis | Coronavirus nsp12-nsp7-nsp8 |
| cis-Acting Elements | Replication initiation sites | PepMV hp1/hp2 stem-loops |
| Template-Switching Signals | Subgenomic RNA synthesis | Coronavirus TRS-B motifs |
| Host Factors | Stabilize replication complexes | hnRNPs, DDX helicases |
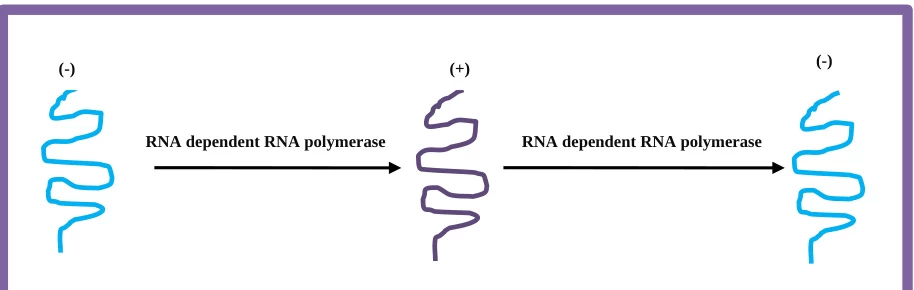
IV. Structural Dynamics of Replication Intermediates
A. Replication Complex Architecture
- Membrane Scaffolding: ROs concentrate replicase components while shielding dsRNA from immune sensors
- Asymmetric Replication: (-)RNA templates produce 10-100x more (+)RNA progeny
B. RNA Secondary Structures
- Pseudoknots: Maintain RdRp processivity during elongation (e.g., flavivirus 3’SL2)
- Slippery Sequences: Facilitate template switching at TRS sites
(Fig. 3: RNA Replication Intermediate)
Description: Molecular view of dsRNA replication complex. Negative-strand (green) base-paired with +ssRNA template (blue). RdRp (red) with nascent RNA chain (yellow).
V. Regulation of Negative-Strand Synthesis
Temporal Control Mechanisms
- Proteolytic Activation: Viral proteases cleave replicase precursors into functional complexes
- Phosphorylation Switches: Host kinases regulate RdRp activity
- RNA Modifications: N6-methyladenosine modulates template accessibility
Strand-Specific Fidelity
- Error Rate: ~10⁻⁴ mutations/base due to lack of proofreading
- Evolutionary Advantage: Facilitates rapid host adaptation and immune evasion
VI. Therapeutic Targeting Opportunities
Inhibition Strategies
| Target | Inhibitor Class | Mechanism |
|---|---|---|
| RdRp Active Site | Nucleotide analogs (Remdesivir) | Chain termination |
| Template Recognition | Aptamers | Block cis-element binding |
| RO Formation | Cyclophilin inhibitors (Cyclosporine) | Disrupt membrane scaffolding |
Diagnostic Applications
- Replication Markers: dsRNA-specific antibodies detect active infection
- Antiviral Screening: Reporter-based replicon systems
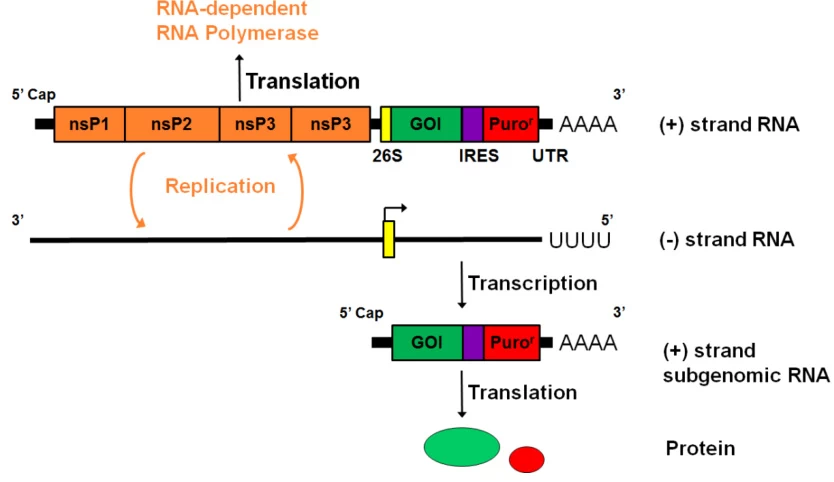
VII. Synthetic Biology Applications
Engineered Replicon Systems
- VEEV-Based Platforms: Self-replicating RNAs for sustained protein expression
- mRNA Vaccine Production: Alphavirus replicons amplifying antigen expression 50x
(Fig. 4: Synthetic Replicon Workflow)
Description: Engineered +ssRNA (blue) with replicase genes (red) and GOI (purple). Continuous amplification via (-)RNA intermediates (green) in transfected cells.
VIII. Unresolved Mechanistic Questions
- How do RdRp complexes maintain processivity across >20kb genomes?
- What triggers the switch from translation to replication?
- Why are (-)RNA synthesis rates strictly limited compared to (+)RNA production?
“Positive-strand RNA viruses exemplify genomic economy: a single molecule serves as infectious agent, mRNA template, and replication blueprint—rewriting central dogma constraints.”
— Cell, 2024
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
