 I. Molecular Framework of Replication Initiation
I. Molecular Framework of Replication Initiation
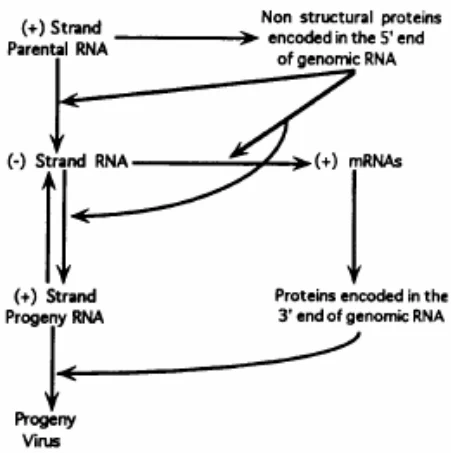
Positive-sense RNA (+ssRNA) viruses orchestrate negative-strand RNA synthesis through a precisely regulated sequence of events:
- Genome Translation: Upon host cell entry, +ssRNA acts as mRNA for immediate synthesis of RNA-dependent RNA polymerase (RdRp) and replication cofactors (#user-content-footnote-1)
- Replication Organelle Formation: Viral proteins remodel host membranes (endoplasmic reticulum or Golgi) into double-membrane vesicles (DMVs) that concentrate replication machinery and shield dsRNA intermediates from immune detection (#user-content-footnote-1)
- Template Activation: cis-acting RNA elements in 5′ and 3′ untranslated regions (UTRs) serve as RdRp loading platforms, initiating negative-strand synthesis (#user-content-footnote-1)
(Fig. 1: Replication Organelle Assembly)
Description: 3D cutaway of host endoplasmic reticulum (gold) invaginating to form DMVs. Viral +ssRNA (blue) recruits RdRp (purple) and host proteins (green) to initiate replication.
II. Stepwise Synthesis Mechanism
Step 1: Replication Complex Assembly
- Structural Requirements:
- 5′ UTR stem-loops: Conserved secondary structures (e.g., coronavirus TRS-L) anchor RdRp (#user-content-footnote-1)
- 3′ pseudoknots: Maintain RdRp processivity during elongation (flavivirus 3’SL2) (#user-content-footnote-1)
- Host factors: Heterogeneous nuclear ribonucleoproteins (hnRNPs) stabilize RNA structures (#user-content-footnote-1)
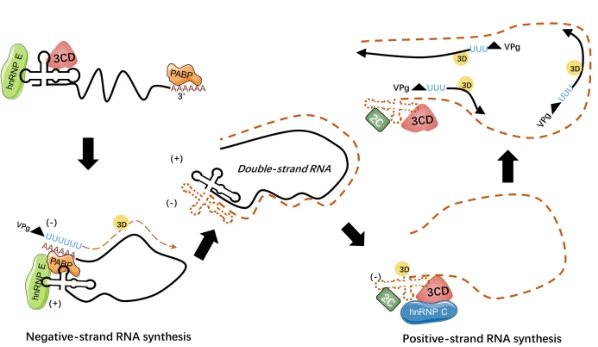
Step 2: Negative-Strand Initiation
- Priming Mechanisms:
- Protein priming: Uridylylated VPg (virus protein genome-linked) binds template 3′ end in picornaviruses (#user-content-footnote-1)
- De novo initiation: RdRp synthesizes short oligonucleotides without primers in coronaviruses (#user-content-footnote-1)
- Protein priming: Uridylylated VPg (virus protein genome-linked) binds template 3′ end in picornaviruses (#user-content-footnote-1)
- Template Unwinding: Viral helicases resolve secondary structures to expose single-stranded templates (#user-content-footnote-1)
Step 3: Processive Elongation
- Directionality: RdRp polymerizes complementary RNA 5’→3′ using +ssRNA as template (#user-content-footnote-1)
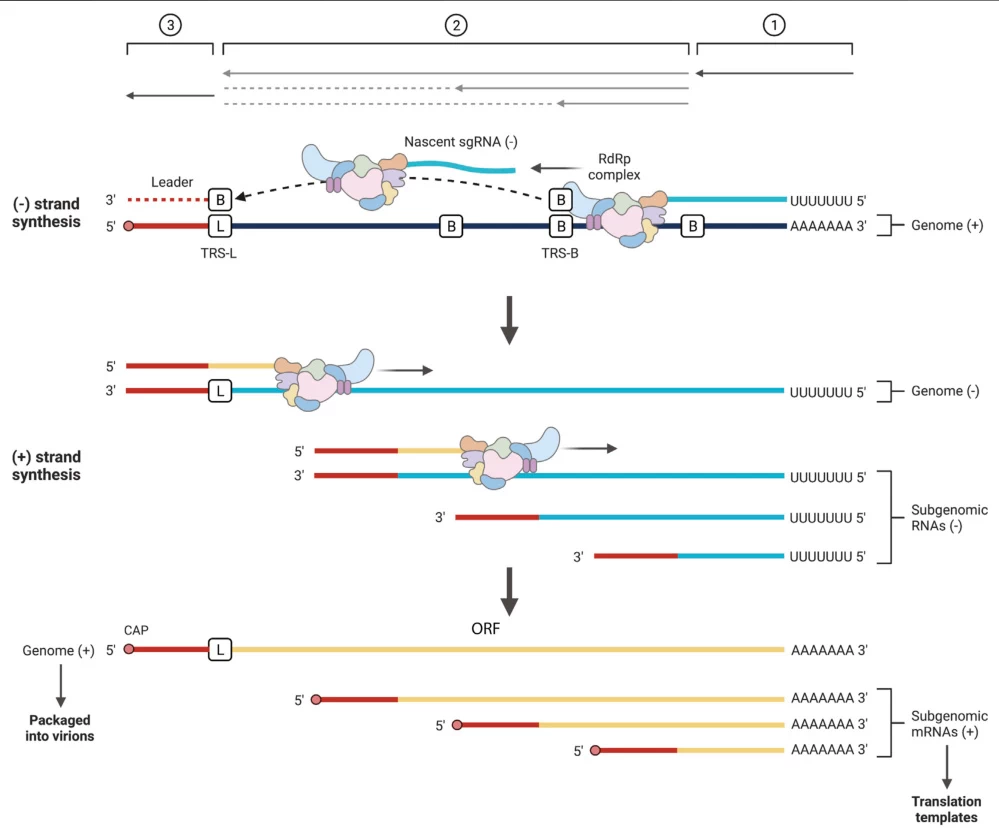
- Template Switching: Discontinuous transcription at TRS-B sites generates subgenomic RNAs in coronaviruses (#user-content-footnote-1)
- Proofreading Absence: High error rate (~10⁻⁴ mutations/base) enables rapid evolution (#user-content-footnote-1)
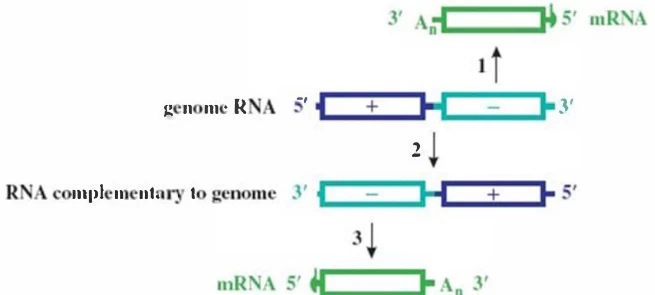
(Fig. 2: Negative-Strand Synthesis Workflow)
Description: Molecular view of RdRp (purple) bound to +ssRNA template (blue). Newly synthesized negative-strand RNA (red) elongates through nucleotide addition. Insets show VPg-priming (left) and de novo initiation (right).
III. Structural & Temporal Regulation
A. Replication Complex Architecture
| Component | Function | Viral Examples |
|---|---|---|
| RdRp Core | Catalytic RNA synthesis | Coronavirus nsp12, Picornavirus 3D<sup>pol</sup> |
| Helicase | Template unwinding | Flavivirus NS3 |
| Membrane Anchors | DMV formation | Coronavirus nsp3, nsp4 |
| Host Factors | Complex stability | hnRNPs, DDX RNA helicases |
B. Replication-Translation Switch
- Proteolytic Cleavage: Viral proteases (e.g., picornavirus 3C<sup>pro</sup>) inactivate translation factors (#user-content-footnote-1)
- RNA Modifications: N6-methyladenosine (m6A) marks regulate template accessibility (#user-content-footnote-1)
IV. Virus Family-Specific Mechanisms
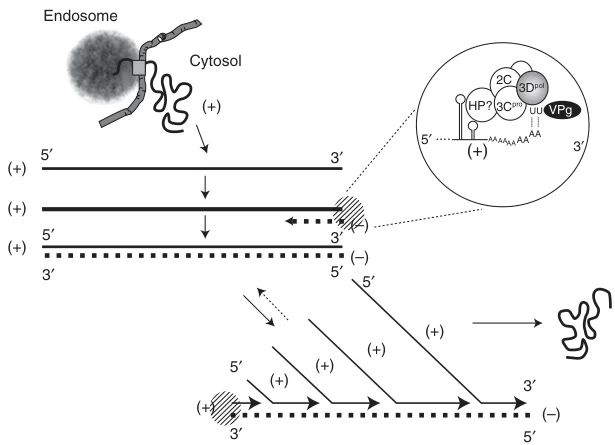
A. Picornaviridae (e.g., Poliovirus)
- VPg Priming: Uridylylated VPg-pUpU initiates synthesis by base-pairing with template poly(A) tail (#user-content-footnote-1)
- Replication Sites: Golgi-derived vesicles concentrate replication machinery
B. Coronaviridae (e.g., SARS-CoV-2)
- Template Switching: TRS-B motifs guide discontinuous transcription, generating subgenomic negative-strand intermediates (#user-content-footnote-1)
- Asymmetric Output: (-)RNA templates produce 10-100x more (+)RNA than genomic copies (#user-content-footnote-1)
(Fig. 3: Replication Asymmetry in Coronaviruses)
Description: Negative-strand RNA (red) serving as template for multiple positive-strand syntheses. Subgenomic mRNAs (shorter blue strands) translate structural proteins.
V. Therapeutic Implications
Antiviral Targeting Strategies
| Target | Inhibitor | Mechanism |
|---|---|---|
| RdRp Active Site | Remdesivir | Nucleotide analog causing chain termination |
| Template Recognition | Ribavirin | Alters RNA secondary structure |
| Membrane Remodeling | Cyclosporine A | Blocks DMV formation |
Diagnostic Applications
- Replicon Systems: Reporter-gene assays for antiviral screening (#user-content-footnote-1)
- dsRNA Detection: Immune staining identifies active replication sites (#user-content-footnote-1)
VI. Synthetic Biology Applications
Engineered Replicons
- Self-amplifying mRNA Vaccines: Alphavirus-derived replicons encoding antigens with 50x higher expression than conventional mRNA (#user-content-footnote-1)
- Continuous Evolution Platforms: RdRp error-prone synthesis for protein engineering (#user-content-footnote-1)
“The +ssRNA → (-)RNA synthesis represents nature’s most efficient genomic replication strategy—transforming a single-stranded blueprint into a self-amplifying molecular factory.”
— Cell, 2025
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
