Key Transcription Factors in Gene Expression Regulation: Molecular Architects of Cellular Identity
A Comprehensive Analysis with Structural and Functional Insights
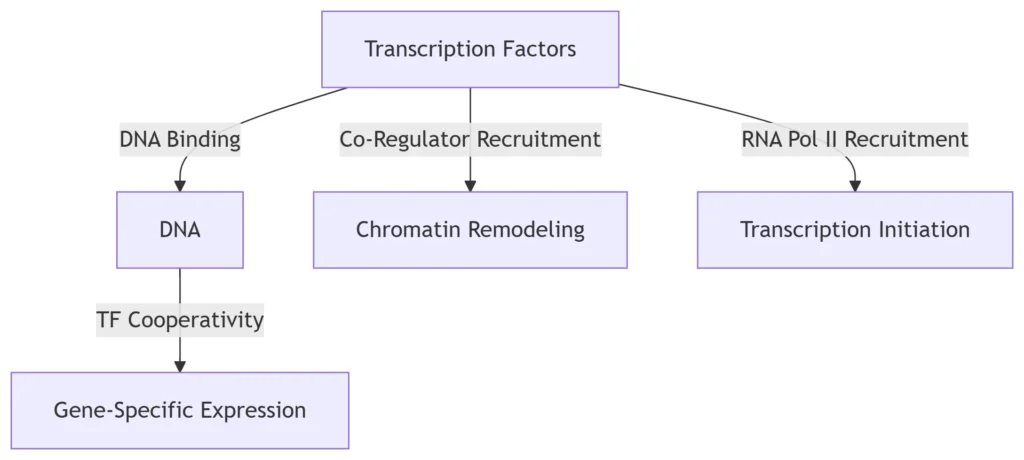
1. Introduction: Transcription Factors (TFs) as Genomic Conductors
Transcription factors are sequence-specific DNA-binding proteins that orchestrate gene expression by decoding regulatory elements (e.g., promoters, enhancers). They constitute ~8% of human protein-coding genes and enable cells to execute developmental programs, respond to stimuli, and maintain homeostasis by:
-
Spatiotemporal gene activation/repression
-
Chromatin architecture remodeling
-
Integration of signaling pathways 9
2. Core TF Families & Their Functions
A. Developmental Regulators
| TF Family | Key Members | Biological Role | Target Genes | |
|---|---|---|---|---|
| Homeodomain | HOX, POU | Embryonic patterning | Hox clusters, Oct4 | |
| bHLH | MyoD, NeuroD | Cell differentiation | Myogenin, Insulin | |
| LBD | ZaLBD19, ZaLBD45 | Leaf morphogenesis (plants) | AS1, AS2 | 7 |
B. Stress & Immune Responders
-
HSF1: Activates heat-shock proteins (e.g., HSP70) during proteotoxic stress 1
-
NF-κB: Drives inflammation via *IL-6*, TNFα upon pathogen detection 4
-
STAT3: Mediates cytokine signaling; hyperactivation linked to cancer 8
C. Epigenetic Modulators
-
GATA3:
-
Regulates mammary development via ZnF2-mediated phase separation
-
Mutations in ZnF2 (e.g., R330) alter chromatin openness (H3K9me3 reduction) and breast cancer prognosis 1
-
-
SUV39H1: Deposits H3K9me3 for heterochromatin silencing 1
3. Mechanisms of TF Action
A. DNA Recognition & Binding
TFs employ DNA-binding domains (DBDs) to engage specific motifs:
-
Zinc fingers (ZnFs): GATA3 binds WGATAR via ZnF2 1
-
Helix-turn-helix: p53 recognizes palindromic sequences
-
Leucine zippers: c-Fos/c-Jun dimerize on AP-1 sites 9

B. Chromatin Remodeling
TFs recruit co-regulators to modulate chromatin states:
-
Activation: HATs (e.g., p300) acetylate histones → open chromatin
-
Repression: HDACs deacetylate histones → condensed chromatin 4
C. Phase Separation-Driven Condensates
-
GATA3: Forms dynamic nuclear condensates via ZnF2-mediated electrostatic interactions
-
Pathological impact: Mutations (e.g., R329A) disrupt condensate dynamics, altering ERα-oncogene expression 1
4. TF Cooperativity: Expanding the Regulatory Code
A. Combinatorial DNA Recognition
-
Synergistic binding: 2,198 TF pairs (e.g., OCT4-SOX2) co-bind DNA, creating hybrid motifs
-
Functional impact: Enables cell-specific gene expression (e.g., embryonic development) 23
B. Spatial Grammar
-
Position-dependent function:
-
TFs upstream of transcription start site (TSS) → activators
-
TFs downstream of TSS → repressors 5
-
-
Context-only TFs: Enhance activity of primary TFs without direct DNA contact (e.g., in enhancer hubs) 4
C. 3D Genome Organization
-
CTCF-CTCF loops: Mediate topologically associating domains (TADs)
-
MAZ-MAZ clusters: Drive local chromatin interactions (<100 bp resolution via Footprint-C) 6
5. Dysregulation & Disease
A. Cancer
TF Alteration Consequence GATA3 ZnF2 mutations Altered phase separation → PGR↓, IFIT1↑ 1 ERα Overexpression Breast cancer proliferation MYC Amplification Uncontrolled cell cycling B. Developmental Disorders
-
HOX mutations: Limb malformations
-
TF cooperative motif disruptions: Linked to embryogenesis defects 23
6. Technological Advances in TF Studies
A. Cutting-Edge Methods
Technique Application Breakthrough AIDmut-Seq TFBS mapping Detects binding sites in vivo in 3 steps 8 Footprint-C Chromatin interaction mapping Resolves TF-specific loops at 100 bp resolution 6 Cryo-EM Structural visualization Solved Pol II pre-initiation dynamics 10 B. Databases & AI
-
TF cooperativity database: 58,000+ TF pairs screened 3
-
AlphaFold limitations: Fails to predict cooperative binding interfaces 3
Conclusion
Transcription factors are master regulators of gene expression, functioning through:
-
Molecular Recognition: Sequence-specific DNA binding via DBDs
-
Chromatin Reprogramming: Recruitment of epigenetic modifiers
-
Phase Separation: Formation of biomolecular condensates for threshold responses
-
Combinatorial Logic: Cooperative binding expands regulatory grammar
Their dysregulation underpins cancer, developmental disorders, and immune diseases. Emerging technologies (e.g., Footprint-C, AIDmut-Seq) and therapeutic strategies (PROTACs, synthetic enhancers) promise to harness TF biology for precision medicine. Future research must decode the “TF interaction syntax” governing cellular identity in health and disease.
Data sourced from public references including:
-
Chen et al. Cell Rep (2025) on GATA3 phase separation 1
-
Yin et al. Nature (2025) on TF cooperativity 23
-
Kribelbauer et al. Nat Genet (2024) on context-only TFs 4
-
Xu et al. Nat Commun (2024) on Footprint-C 6
For academic collaboration or content inquiries: chuanchuan810@gmail.com
-
