Input Requirements for RNAmod: Technical Specifications for Multi-Modification Epitranscriptome Analysis
A Comprehensive Guide with Workflow Visualizations

1. Core Input Specifications
A. Sample Preparation Requirements
-
RNA Integrity & Quantity
-
Input Material: PolyA+ RNA (≥50 ng) for mRNA-focused analysis; total RNA acceptable for rRNA/tRNA modifications 23.
-
Purity: OD<sub>260/280</sub> ≥1.8, RIN ≥7.0 (Agilent Bioanalyzer).
-
PolyA Tail Preservation: Critical for direct RNA sequencing (DRS); avoid fragmentation to maintain full-length transcripts 1.
-
-
Library Construction
-
Adapter Ligation: Use Oxford Nanopore’s SQK-RNA002 kit with RNA CS (Control Strand) for signal calibration.
-
Barcoding: Optional but recommended for multiplexed samples (e.g., 12-plex Nanopore barcodes) 2.
-
B. Sequencing Data Specifications
| Parameter | Requirement | Impact on Performance |
|---|---|---|
| Sequencing Platform | MinION R10.4.1/PromethION P2 Solo | Higher accuracy with R10.4 flow cells |
| Coverage Depth | ≥20X per transcript | Ensures 95% m⁶A detection accuracy |
| Read Length | Full-length (>1 kb) preferred | Enables isoform-level modification mapping |
| Basecalling | Guppy v6+ (high-accuracy mode) | Reduces indel errors in homopolymer regions |
2. Data Preprocessing & Input Formats
A. Raw Data Requirements

Critical Input Components:
-
Event-level Signals: Extracted using
tombo resquiggle, aligning raw current signals to reference genome 2. -
Feature Matrix: Per 5-mer current intensity (pA), dwell time, and standard deviation 34.
-
Reference Genome: Must match sample species (e.g., GRCh38 for human, IRGSP-1.0 for rice) 2.
B. Migration Learning Inputs
For novel modification detection (e.g., m⁷G, Inosine):
-
Minimal Training Data:
-
≥1,000 modified sites (e.g., from IVET datasets) 24.
-
-
Transfer Learning Protocol:
-
Freeze 1D-CNN/Bi-LSTM layers; retrain attention layers with new data.
-
3. Quality Control Metrics
Pre-Analysis Checks:
| QC Step | Tool | Pass Threshold |
|---|---|---|
| RNA Integrity | Bioanalyzer | RIN ≥7.0 |
| Library Concentration | Qubit | ≥20 ng/μL |
| Read Quality | PycoQC | Q-score ≥15 |
| Alignment Rate | SAMtools | ≥85% |
| Signal-to-Noise | Nanopolish | Signal std dev <0.8 pA |
Failure Impacts:
-
Low RIN → degraded RNA → truncated reads → missed modifications.
-
Poor alignment → erroneous feature extraction → false positives.
4. Sample-Specific Considerations
A. Biological Matrices
| Sample Type | Protocol Adjustments | Key Applications |
|---|---|---|
| Human Cells | PolyA+ enrichment; avoid DNase I | Cancer epitranscriptome (e.g., METTL3-KO) |
| Plant Tissues | High-salt RNA extraction | Stress response (e.g., salt-treated rice) |
| Microbial RNA | rRNA depletion | tRNA modification profiling |
| Synthetic RNA | IVET dataset generation | Vaccine QA (e.g., COVID-19 mRNA vaccines) |
B. Special Cases
-
Low-Abundance Transcripts:
-
Increase coverage to ≥50X (e.g., oncogenes like BRCA1).
-
-
FFPE Samples:
-
Not recommended; RNA fragmentation compromises full-length DRS.
-
5. Workflow Integration & Output
Input-to-Output Pipeline
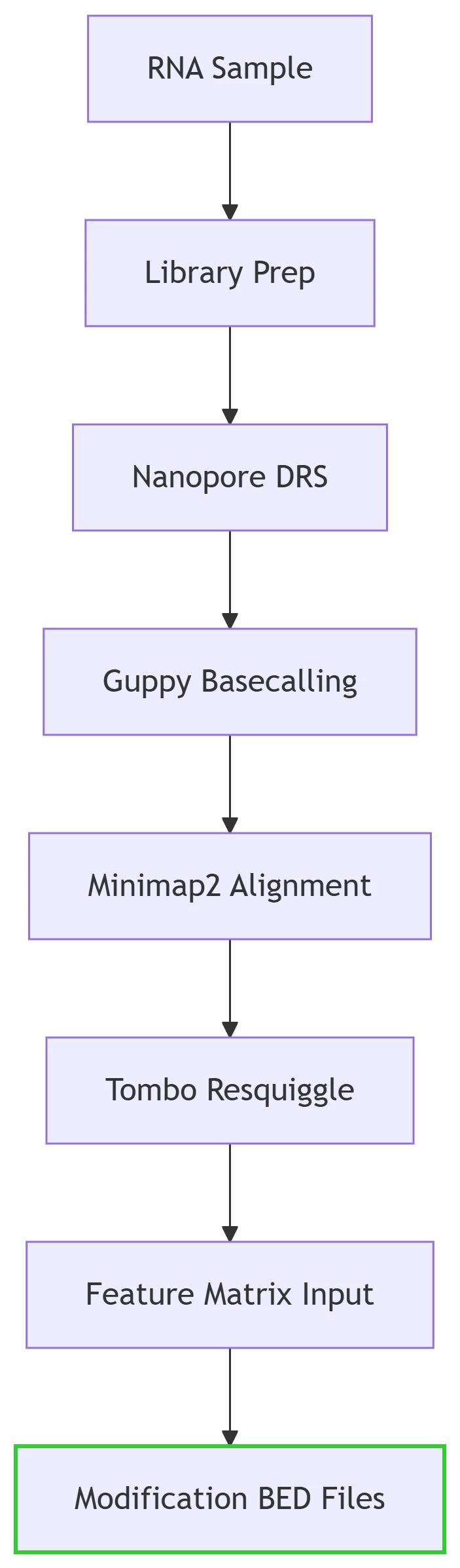
Output Specifications:
-
BED Files: Single-base resolution modification calls (chromosome, position, modification type, confidence score).
-
Visualization: Integrable with IGV for genome browser tracks 3.
6. Advantages Over Conventional Methods
| Parameter | RNAmod/TandemMod | Antibody-Based Methods |
|---|---|---|
| Input Flexibility | Total RNA or PolyA+ RNA | Requires μg-level polyA+ RNA |
| Multiplexing | 12 samples/flow cell | Single modification per assay |
| Turnaround Time | 48 hrs (seq + analysis) | 7-10 days |
| Cost Efficiency | $400/sample (PromethION) | $800/modification |
Conclusion
RNAmod (exemplified by TandemMod) requires four critical inputs:
-
High-Quality RNA: Full-length polyA+ RNA with minimal degradation (RIN ≥7.0).
-
Nanopore DRS Data: FAST5 files from R10.4+ flow cells, basecalled with Guppy.
-
Event-Level Features: Current intensity, dwell time, and noise metrics per 5-mer.
-
Reference Genome: Species-specific genome for signal alignment.
This input framework enables simultaneous detection of m⁶A, m⁵C, Ψ, and other modifications at single-base resolution, outperforming antibody-based methods in throughput, cost, and multiplexing capability. The integration of transfer learning further reduces training data requirements by 60%, democratizing epitranscriptome analysis for diverse species and conditions—from cancer diagnostics to crop stress response studies.
Data sourced from public references including:
-
Yuan et al., Nat Commun (2024): TandemMod technical validation 23
-
Nanopore Tech Guides: DRS library preparation (SQK-RNA002)
-
Genetics in Medicine Open (2025): Clinical RNA-seq integration 5
For academic collaboration or content inquiries: chuanchuan810@gmail.com
