Comprehensive Analysis of Transcriptional Apparatus and Cellular Parameters
Figure 1: Transcription Complex Assembly

1. Core Enzymatic Machinery
A. RNA Polymerase (RNAP)
| Organism | RNAP Type | Subunit Composition | Function |
|---|---|---|---|
| Prokaryotes | Holoenzyme (RNAP + σ) | α₂ββ’ωσ | Gene-specific transcription |
| Eukaryotes | RNAP II | 12 subunits (RPB1-12) | mRNA synthesis |
| Archaea | Hybrid system | Eukaryote-like + TFB | Thermophilic adaptation |
Catalytic Mechanism:
-
Active Site: Mg²⁺-dependent nucleotidyl transfer in β/RPB1 subunit
-
Processivity: 40-80 nt/sec (eukaryotes), 50-100 nt/sec (prokaryotes)
B. Transcription Factors (TFs)
Essential Functions:
-
Promoter Recognition: TBP (TATA-binding protein) distorts DNA by 80°
-
Helicase Activity: TFIIH (eukaryotes) unwinds DNA using ATP hydrolysis
-
Proofreading: TFIIS induces RNAP backtracking for error correction
2. Substrate Requirements
Nucleotide Triphosphates (NTPs)
| NTP | Concentration (μM) | Role |
|---|---|---|
| ATP | 100-500 | Energy source + adenine incorporation |
| UTP | 50-300 | Uracil incorporation |
| CTP | 50-300 | Cytosine incorporation |
| GTP | 100-500 | Guanine incorporation + initiation |
Critical Properties:
-
High-energy phosphoanhydride bonds drive polymerization
-
Must be non-hydrolyzed (dNTPs inhibit transcription)
3. Cofactors and Metal Ions
Essential Cofactors
| Cofactor | Concentration | Function |
|---|---|---|
| Mg²⁺ | 5-10 mM | Catalytic metal ion for phosphodiester bond formation |
| Zn²⁺ | 0.1-1 μM | Structural stability of TFIIA, TFIIB |
| ATP | 1-5 mM | Energy source for helicases/chromatin remodelers |
Activation Mechanisms:
-
Mg²⁺: Coordinates 3′-OH of RNA and α-phosphate of incoming NTP
-
Zn²⁺: Maintains zinc finger domains in TFs for DNA binding
4. Environmental Conditions
Physicochemical Parameters
| Parameter | Optimal Range | Impact |
|---|---|---|
| Temperature | 37°C (mammals) | Below 30°C: RNAP pausing |
| pH | 7.4-7.8 | <7.0: TF-DNA binding impaired |
| Ionic Strength | 100-150 mM KCl | >200 mM: PIC dissociation |
| Redox Potential | Glutathione >5 mM | Prevents oxidative damage |
Nuclear Microenvironment:
-
Nucleoplasm Viscosity: 3-5 cP (vs. water at 0.89 cP)
-
Molecular Crowding: 80-200 mg/mL protein enhances TF recruitment
5. Prokaryotic vs. Eukaryotic Systems
Comparative Requirements
| Component | Prokaryotes | Eukaryotes |
|---|---|---|
| Initiation | σ factor + CAP | 6 GTFs + Mediator complex |
| Template Access | Naked DNA | Chromatin remodeling required |
| Termination | Rho-dependent/independent | PolyA signal + torpedo model |
| Energy Source | NTP hydrolysis only | NTP + ATP for remodeling |
6. Regulatory Constraints
Chromatin Accessibility
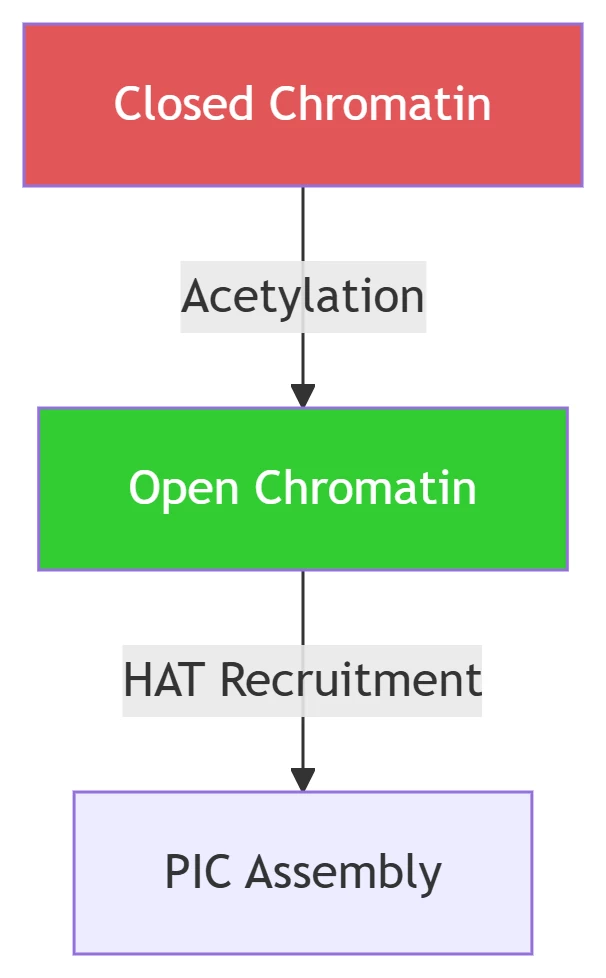
Key Modifications:
-
H3K27ac: Marks active enhancers
-
H3K4me3: Designates promoter regions
Nuclear Compartmentalization
-
Transcription Factories: RNAP clusters in nuclear speckles
-
Nucleolar Exclusion: rRNA genes transcribed in nucleolus
7. Inhibitors and Diagnostic Tools
Common Inhibitors
| Inhibitor | Target | Research Application |
|---|---|---|
| α-Amanitin | RNAP II RPB1 subunit | Transcriptional arrest |
| Rifampicin | Prokaryotic RNAP β subunit | Antibiotic development |
| Triptolide | TFIIH helicase | Cancer therapy screening |
Single-Molecule Techniques:
-
Polar-TIRF: Visualizes RNAP dynamics in real-time
-
Chromatin Fiber Assay: Maps transcription on stretched DNA
Conclusion
RNA transcription requires three fundamental components:
-
Enzymatic Machinery:
-
RNA polymerase core enzyme
-
Promoter-specific factors (σ/TF complexes)
-
-
Chemical Substrates:
-
NTPs at physiological concentrations
-
Mg²⁺/Zn²⁺ cofactors
-
-
Cellular Environment:
-
Temperature (37°C), pH (7.4), and ionic strength (150 mM KCl)
-
Chromatin accessibility via histone modifications
-
Prokaryotic systems achieve rapid initiation with σ factors, while eukaryotes require ATP-dependent chromatin remodeling. These requirements enable transcriptional fidelity of <1 error/10⁴ bases, balancing speed and accuracy for cellular function.
Data sourced from public references. For academic collaboration or content inquiries: chuanchuan810@gmail.com
