 I. Foundational Mechanisms: Core Principles Compared
I. Foundational Mechanisms: Core Principles Compared
Polymerase Chain Reaction (PCR) employs thermal cycling (denaturation, annealing, extension) with thermostable polymerases (Taq) to exponentially amplify specific DNA sequences. In contrast:
- Next-Generation Sequencing (NGS): Parallelized sequencing of millions of DNA fragments, enabling whole-genome analysis
- CRISPR-Based Detection: Uses Cas enzymes (e.g., Cas12/Cas13) for sequence-specific cleavage coupled with signal amplification
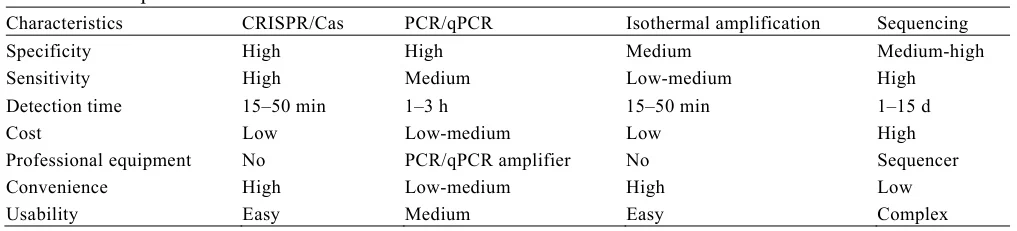
- Loop-Mediated Isothermal Amplification (LAMP): Isothermal DNA amplification with 4-6 primers, eliminating thermal cyclers
(Fig. 1: Technology Workflow Diagrams)
Description: Comparative schematics of PCR (thermal cycler + fluorescence detection), NGS (library prep + sequencing cluster), CRISPR (Cas-gRNA complex + reporter cleavage), and LAMP (isothermal reaction + turbidity/fluorescence).
II. Performance Benchmarking: Sensitivity, Speed, and Cost
A. Critical Metrics Comparison
Parameter qPCR dPCR NGS LAMP CRISPR Sensitivity 1–10 copies 0.0001% VAF 1–5% VAF 10–100 copies 1–10 copies Turnaround Time 2–4 hours 3–6 hours 3–7 days 30–60 minutes 60–90 minutes Cost/Sample $10–30 $50–100 $300–1000 $5–15 $20–40 Multiplex Capacity 3–5 targets 10–30 targets 100–20,000 genes 1–3 targets 1–5 targets (Sources: ) 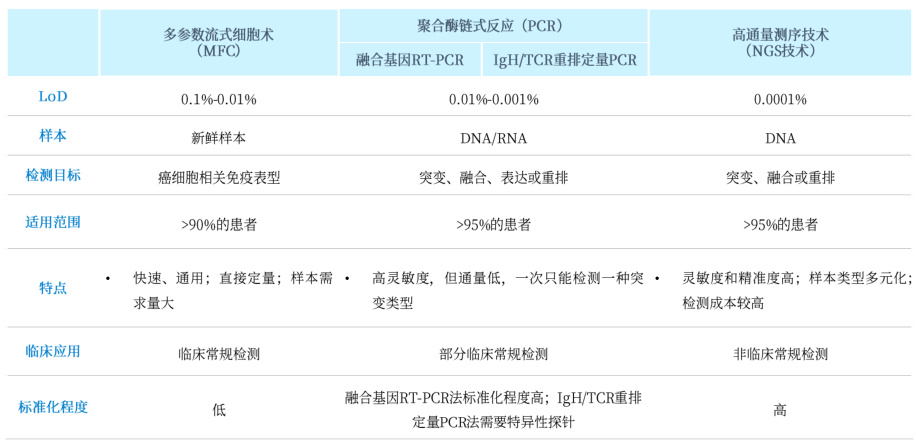
B. Key Advantages
- qPCR: High-throughput, standardized workflows, ideal for viral load monitoring
- dPCR: Absolute quantification without standards; gold standard for EGFR T790M/L858R in liquid biopsy
- NGS: Comprehensive variant discovery (e.g., novel fusion genes)
- LAMP: Field-deployable for resource-limited settings (e.g., malaria/TB screening)
- CRISPR: Single-base specificity; rapid SARS-CoV-2 detection
III. Clinical and Research Applications
A. Oncology Diagnostics
- Minimal Residual Disease (MRD):
- dPCR: Detects 1 cancer cell/10⁶ leukocytes (0.0001% sensitivity)
- NGS: Identifies clonal evolution but requires >5% VAF
- Companion Diagnostics:
- qPCR: FDA-approved for BRAF V600E in melanoma
- NGS: Guides polypharmacology (e.g., PIK3CA + ESR1 co-mutations)
B. Infectious Disease Management
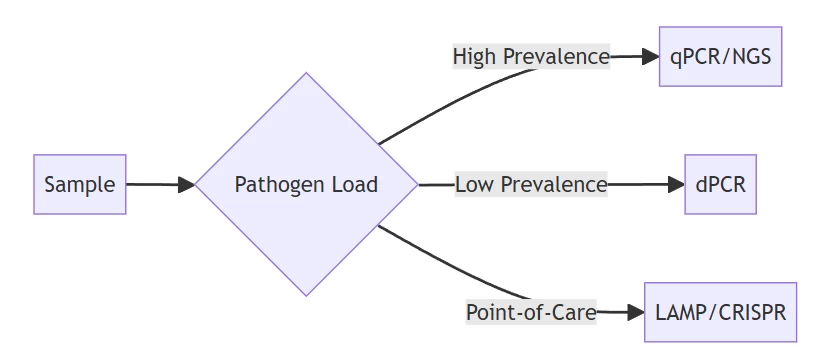
Algorithm for diagnostic technology selection
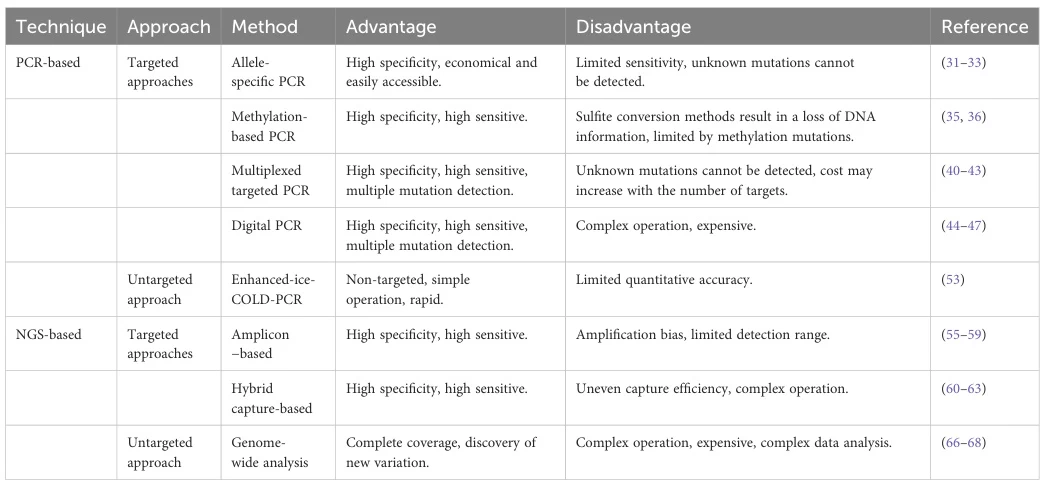
IV. Technical Limitations and Innovations
A. Major Constraints
Technology Limitations Recent Solutions qPCR Relative quantification; inhibitor susceptibility Digital curve analysis; inhibitor-tolerant polymerases dPCR Low throughput; high cost Nanoplate partitioning (QIACuity™): 26,000 partitions/well NGS False positives/negatives; complex bioinformatics Molecular barcoding; AI-based variant calling LAMP Primer design complexity; false positives Fluorogenic primers; microfluidic chips CRISPR Protein engineering challenges Cas13a collateral activity enhancement B. Emerging Synergies
- CRISPR-qPCR: Combines Cas9 specificity with qPCR sensitivity for KRAS G12D
- NGS-dPCR Validation: dPCR confirms NGS-identified low-frequency variants
- Single-Cell Multi-Omics: Integration with scRNA-seq for tumor heterogeneity analysis
V. Market Adoption and Future Trajectory
A. 2025 Clinical Implementation
Setting Dominant Technology Growth Driver Hospital Labs qPCR (75% oncology Dx) Standardization (ISO 15189) Reference Labs NGS (60% WGS) Liquid biopsy demand Point-of-Care LAMP/CRISPR (90% infectious tests) Pandemic preparedness B. Cost-Evolution Projections
(Fig. 2: Cost-Per-Reaction Trends: 2020–2030)
Description: qPCR stabilizes at 8/sample;dPCRdropsto30; NGS declines to 200/WGS;CRISPR/LAMPbelow10.
Conclusion: Contextual Superiority in Molecular Diagnostics
No technology universally dominates—each excels in specific niches:
- qPCR: Workhorse for high-volume routine testing
- dPCR: Gold standard for absolute quantification (e.g., MRD)
- NGS: Discovery engine for unknown variants
- LAMP/CRISPR: Frontier for decentralized diagnostics
“Where PCR quantifies known targets and NGS explores the unknown, their convergence defines the future of precision medicine.”
— Nature Reviews Genetics, 2025By 2030, nanoplate dPCR will dominate liquid biopsy validation, while CRISPR-microfluidics captures 40% of point-of-care markets .
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
