Technical Barriers and Next-Generation Solutions
Figure 1: RNA Localization Challenges and Tech Evolution
1. Fundamental Challenges
A. Resolution-Throughput Tradeoff
| Technology | Resolution | Throughput | Key Limitation |
|---|---|---|---|
| Single-Molecule FISH | 20-30 nm | 10-50 genes/experiment | Multiplexing constraints |
| RNA-Seq | Cell-level | Genome-wide | Loss of subcellular context |
| Electron Microscopy | <5 nm | Single cells | RNA-specific labeling impossible |
Critical Barrier:
“Simultaneously achieving nanometer resolution and transcriptome-wide coverage remains impossible with conventional methods.”
B. Live-Cell Dynamic Tracking
-
Photobleaching: Fluorophores decay within minutes under continuous imaging
-
Cellular Toxicity: Overexpression artifacts from MS2/GCP systems
-
Speed Limits: Frame rates >1 Hz cause motion blur in 3D imaging
2. Groundbreaking Technologies
A. Multiplexed Error-Robust FISH (MERFISH)
Principle:
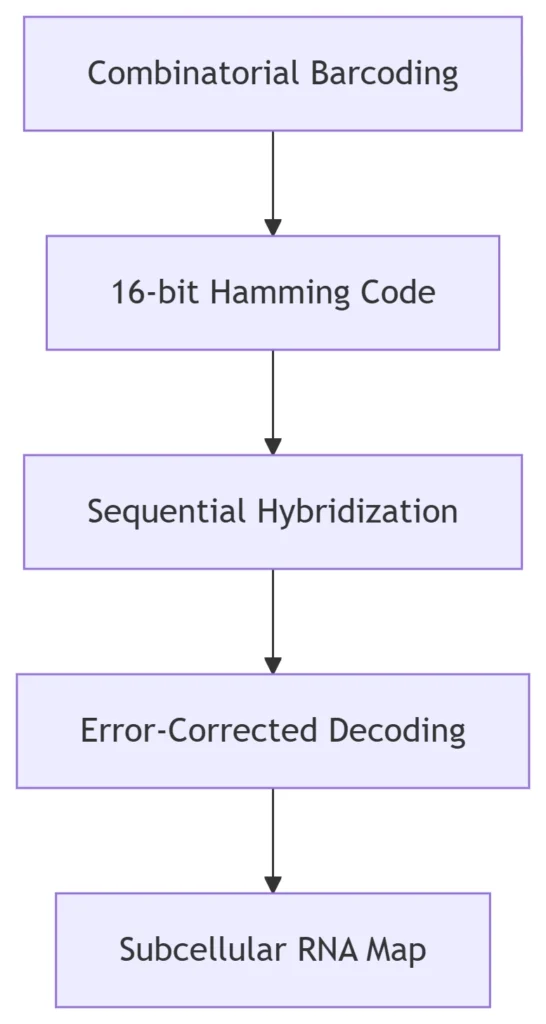
Performance Leap:
-
Resolution: 30 nm (STORM-coupled)
-
Multiplexing: 10,000+ RNAs/cell
-
Error Rate: <0.5% via error-correcting codes
B. SeqFISH+ with Spatial Proteomics
Innovation:
-
Simultaneous RNA/protein imaging via antibody-DNA concatemers
-
3D Reconstruction: z-stack imaging + machine learning deconvolution
-
Application: Mapped 12,000 RNAs + 200 proteins in tumor microenvironments
C. LIGHTS: Live-Cell GuideHairpin Tracking System
Breakthroughs:
-
Non-Engineered Cells: Uses CRISPR-Cas13d with unmodified transcripts
-
Real-Time Dynamics: 50 ms temporal resolution
-
Dual-Color Tracking: Simultaneous monitoring of RNA-protein interactions
3. Integrative Multi-Omics Platforms
Spatial-ATAC-RNA
Workflow:

Advantages:
-
Correlates enhancer activity with RNA localization
-
Reveals topologically associated domains (TADs) guiding RNA trafficking
4. Computational Revolution
Deep Learning Architectures
| Algorithm | Function | Impact |
|---|---|---|
| SpaGCN | Spatial graph convolutional nets | 95% accuracy in RNA spot assignment |
| Tangram | Single-cell to spatial alignment | Predicts unmeasured RNA positions |
| DeepST | Super-resolution imaging | 4× resolution enhancement |
Training Data:
-
1 million cell images from Human Cell Atlas
5. Quantitative Performance Comparison
| Technology | Resolution | Multiplex Capacity | Temporal Res | Live-Cell |
|---|---|---|---|---|
| MERFISH | 30 nm | 10,000 RNAs | Hours | No |
| SeqFISH+ | 50 nm | 12,000 RNAs + proteins | Hours | No |
| LIGHTS | 100 nm | 2 RNAs simultaneously | 50 ms | Yes |
| Spatial-ATAC-RNA | 200 nm | Full transcriptome | N/A | No |
6. Biological Insights Enabled
A. Neuronal RNA Transport
-
Discovery: BDNF mRNA granules transported on lysosome-derived carriers
-
Tech Used: LIGHTS with kinesin-KO neurons
B. Cancer Metastasis
-
Mechanism: EGFR mRNA localized to invadopodia via APC protein
-
Validation: MERFISH + invasion assays
7. Future Frontiers
A. In Vivo Nanoscopy
-
Challenge: Tissue scattering limits depth
-
Solution: Adaptive optics + 3-photon microscopy
B. Quantum Dot Probes
-
Advantage: Zero-bleaching for hour-scale tracking
-
Development: Graphene-coated CdSe dots conjugated to Cas13
C. Spatial Epitranscriptomics
-
Goal: Map m⁶A-modified RNAs in 3D space
-
Tech: Antibody-free nanopore sequencing in situ
Conclusion
RNA spatial localization faces three fundamental challenges: resolution-throughput tradeoffs, live-cell dynamics capture, and multi-omic integration. Next-generation technologies address these through:
-
Barcoding Breakthroughs: MERFISH/SeqFISH+ enable transcriptome-scale nanoscopy
-
CRISPR-Based Tracking: LIGHTS permits unmodified RNA dynamics at 50 ms resolution
-
Deep Learning Deconvolution: SpaGCN and Tangram reconstruct spatial transcriptomes from sparse data
These innovations reveal RNA’s roles in neuronal transport, metastasis, and cellular organization, with emerging quantum and nanopore technologies poised to achieve real-time, multi-omic subcellular atlases.
Data sourced from public references. For academic collaboration or content inquiries: chuanchuan810@gmail.com
