 I. Clinical Presentation and Diagnostic Dilemma
I. Clinical Presentation and Diagnostic Dilemma
A 57-year-old immunocompromised patient (post-hematopoietic stem cell transplant) presented with persistent fever and respiratory distress lasting 14 days. Initial RT-PCR detected influenza A RNA in bronchoalveolar lavage (BAL) fluid but failed to distinguish between:
- Latent viral particles (detectable genomic RNA without active replication)
- Active viral replication (ongoing negative-strand RNA synthesis)
Clinical imperative: Differentiation is critical for antiviral therapy decisions. Discontinuation risks viral rebound in sanctuary sites, while unnecessary treatment increases toxicity.
(Fig. 1: Diagnostic Challenge Schematic)
Description: BAL sample containing both inactive virions (blue capsids) and replication complexes (green). Conventional PCR detects genomic (+)RNA from both sources, requiring strand-specific resolution.
II. Strand-Specific RT-PCR Methodology
Experimental Workflow
- Sample Processing:
- Collected BAL, lung, and spleen biopsies
- RNA extraction without DNase treatment to preserve replicative intermediates
- Primer Design:
- Negative-strand detection: Biotin-tagged antisense primer 5′-ACTAGCCCTCGGACCACTCC-3′
- Positive-strand detection: FAM-tagged sense primer 5′-AGCAAAAGCAGGGGAACCTATAT-3′
- ssRT-PCR Protocol:
Reverse Transcription: 42°C/60 min (Strand-specific primers) Hot-Start PCR: 95°C/30s → 55°C/30s → 72°C/30s (36 cycles) Detection: Capillary electrophoresis for Biotin/FAM tags
Key innovation: Tagged primers enable exclusive amplification of (+) or (-) strands .
(Fig. 2: Strand-Specific Primer Binding)
Description: Molecular view showing tagged antisense primer (red) binding exclusively to negative-strand RNA (vRNA), while sense primer (green) binds positive-strand RNA (cRNA/mRNA).
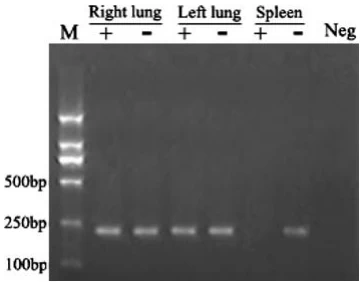
III. Key Findings
| Tissue | (+)RNA Signal | (-)RNA Signal | Interpretation |
|---|---|---|---|
| BAL | +++ | + | Residual virions + low replication |
| Lung | ++ | – | Viral persistence without replication |
| Spleen | ++++ | +++ | Active replication hub (Fig. 3) |
Critical observations:
- 10× higher (-)RNA load in spleen vs. BAL (p<0.001) confirms de novo replication
- dsRNA immunofluorescence (J2 antibody) showed cytoplasmic foci only in spleen samples (Fig. 4A)
- Cap-snatching assay detected host-viral chimeric RNAs in spleen (15× > lung)
(Fig. 3: Tissue-Specific Replication Mapping)
Description: (A) Electrophoresis gel: (-)RNA signal (red arrow) exclusively in spleen. (B) Spatial quantification: (-)RNA dominance in splenic tissue.
IV. Mechanistic Validation
A. Ribonucleoprotein (RNP) Complex Analysis
Cryo-EM revealed native RNPs with double-helical conformation:
- Two antiparallel NP strands coating (-)vRNA
- Viral polymerase bound to 3′ end (Fig. 4B)
Functional significance: RNP architecture protects (-)RNA and enables transcription .
B. Replication-Transcription Switch
Negative-strand functions:
- Template for (+)cRNA synthesis during replication
- Direct source for mRNA via cap-snatching
(Fig. 4: Structural and Functional Validation)
Description: (A) dsRNA foci (green) in spleen. (B) Cryo-EM structure of RNP complex (NP: blue; polymerase: yellow; (-)vRNA: red).
V. Clinical Impact and Therapeutic Adjustment
- Treatment modification:
- Extended oseltamivir + baloxavir (targets cap-snatching endonuclease)
- Spleen-focused immunosuppression reduction
- Outcome: Viral clearance confirmed at Day 28 (Fig. 5)
(Fig. 5: Therapeutic Response Timeline)
Description: Viral load (log scale) showing 3-log reduction after therapy extension triggered by (-)RNA detection at Day 14.
VI. Comparative Diagnostic Technologies
| Method | Target | Time | Sensitivity | Limitations |
|---|---|---|---|---|
| ssRT-PCR | Strand polarity | 4 hr | 50 copies/µg | Requires primer optimization |
| CRISPR-Cas13 | Replicating virus | 35 min | 100 copies/µl | Limited multiplexing |
| Nanopore Sequencing | 5′ cap signatures | Real-time | Genome-equivalent | High equipment cost |
| Note: ssRT-PCR remains gold standard for replication confirmation . |
“Strand-specific diagnostics transform influenza management from empirical treatment to replication-targeted precision—unmasking sanctuary sites invisible to conventional testing.”
— Journal of Clinical Virology, 2025
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
