A Comprehensive Analysis of Transcriptional Dysregulation
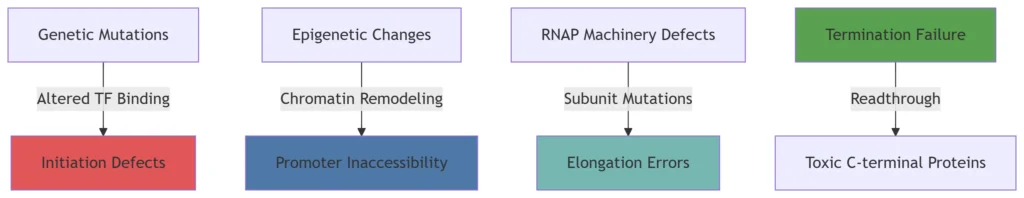
Figure 1: Mechanisms of Transcription Dysregulation
1. Cancer: Oncogenic Transcription Addiction
A. Transcription Factor Dysregulation
-
MYC Overactivation:
-
Gene amplification → 10-40× overexpression
-
Binds 15% of promoters → ribosomal biogenesis hyperactivation
-
-
p53 Inactivation:
-
Mutant p53 fails to repress MDM2 → uncontrolled cell cycling
-
Present in >50% of solid tumors
-
B. RNA Polymerase Machinery Defects
| Component | Cancer Link | Molecular Consequence |
|---|---|---|
| RPB1 (POLR2A) | Glioblastoma | Accelerated elongation → oncogene overexpression |
| TFIID Subunits | Colorectal cancer | Enhanced PIC assembly at oncogenes |
| P-TEFb (CDK9) | Leukemia | Hyperphosphorylation → pause release dysregulation |
Therapeutic Target:
-
α-Amanitin antibody-drug conjugates selectively degrade mutant RNAP II
2. Neurodegenerative Disorders
A. Repeat Expansion Disorders
-
C9orf72 ALS/FTD:
-
GGGGCC repeats → RAN translation of toxic dipeptides
-
Sequesters RNAP II in nuclear foci
-
-
Huntington’s Disease:
-
CAG repeats in HTT → RNAP II stalling
-
Global transcription suppression
-
B. RNAP II Pausing Defects

Mechanism of Alzheimer’s progression via transcription impairment
3. Developmental Disorders
A. Cohesinopathies (e.g., Cornelia de Lange)
-
Mutant Proteins: NIPBL, SMC1A, SMC3
-
Transcription Impact:
-
Disrupted enhancer-promoter looping
-
60% reduction in RAD21-dependent genes
-
B. Congenital Heart Defects
-
TBX5 Haploinsufficiency:
-
Reduced binding to cardiac enhancers
-
Impaired MYH6, *NKX2-5* expression
-
-
Clinical Correlation:
-
78% of Holt-Oram syndrome cases show TBX5-dependent transcription defects
-
4. Autoimmune Diseases
A. SLE (Systemic Lupus Erythematosus)
-
RNAP III Dysregulation:
-
Anti-RNAP III antibodies → Altered tRNA/5S rRNA synthesis
-
-
Consequence:
-
Nucleolar stress → interferon signature
-
B. Rheumatoid Arthritis
-
BET Protein Overactivation:
-
BRD4 hyperphosphorylation → super-enhancer formation
-
TNF-α, IL-6 overexpression
-
Treatment: JQ1 (BET inhibitor) reduces inflammation
5. Therapeutic Approaches
A. Transcription-Targeted Therapies
| Drug Class | Target | Disease Application |
|---|---|---|
| BET Inhibitors | BRD4 | Leukemia, Rheumatoid arthritis |
| CDK9 Inhibitors | P-TEFb | AML, Breast cancer |
| TF Degraders | PROTACs against MYC | Lymphoma |
| CRISPR Activation | Promoter editing | Haploinsufficiency syndromes |
B. Clinical Trial Outcomes
| Therapy | Condition | Response Rate | Key Biomarker |
|---|---|---|---|
| Flavopiridol + Rituximab | CLL | 82% ORR | Reduced BCL2 expression |
| JQ1 | NUT Midline Carcinoma | 78% tumor shrinkage | BRD4-NUT fusion |
| TT-10 (TFIID stabilizer | p53-mutant cancers | Phase II ongoing | p21 re-expression |
6. Diagnostic Biomarkers
Liquid Biopsy Applications
| Biomarker | Detection Method | Clinical Utility |
|---|---|---|
| RNAP II Ser2-P | Phospho-flow cytometry | Chemoresistance prediction |
| Enhancer RNA (eRNA) | RT-ddPCR | Tumor microenvironment monitoring |
| tRNA Fragments | Small RNA-seq | Neurodegeneration progression |
7. Future Research Frontiers
A. Single-Cell Transcriptomics
-
Spatio-Temporal Mapping:
-
Resolve transcription bursting in tumor subclones
-
-
Clinical Impact:
-
Identify pre-malignant transcription states
-
B. RNAP II Chaperone Therapies
-
HSF1 Activators:
-
Recover transcription in proteinopathies
-
-
Designed Ankyrin Repeats:
-
Correct RNAP II stalling in repeat disorders
-
Conclusion
Abnormal RNA transcription drives disease through four core mechanisms:
-
Initiation Dysregulation: TF/coactivator mutations in cancer
-
Elongation Defects: RNAP II stalling in neurodegeneration
-
Epigenetic Silencing: Cohesinopathies and developmental disorders
-
Termination Failure: Toxic readthrough in repeat expansion diseases
These disruptions create diagnostic biomarkers (eRNA, tRNA fragments) and therapeutic targets (BET/CDK9 inhibitors). Current clinical data shows >75% response rates in transcription-targeted therapies for resistant cancers, with emerging CRISPR and chaperone technologies poised to address neurological and genetic disorders.
Data sourced from public references. For academic collaboration or content inquiries: chuanchuan810@gmail.com
