 I. Foundational Workflow Architecture
I. Foundational Workflow Architecture
RNA extraction kits employ systematic phase-separation biochemistry to isolate high-integrity RNA through four universal stages:
- Cellular Disruption & Lysis
- Contaminant Removal & RNA Binding
- Matrix Purification & Washing
- Elution & Quality Verification
(Fig. 1: Universal RNA Extraction Workflow)
Description: Circular diagram with color-coded phases: Tissue homogenization (red), Phase separation (blue), Column purification (green), QC analysis (gold). Arrows indicate directional workflow progression.
II. Phase 1: Lysis & Initial Processing
A. Sample-Specific Disruption Strategies
| Sample Type | Lysis Technology | Critical Reagents |
|---|---|---|
| Plant tissues | Mechanical grinding + Buffer S1 | β-mercaptoethanol, chaotropic salts |
| FFPE samples | Xylene deparaffinization → Proteinase K | Cross-link reversal buffers |
| Exosomes | Immunomagnetic capture → Ethanol precipitation | Anti-exosome antibodies |
| Bacteria | Lysozyme/lysis buffer treatment | TE buffer with chaotropic agents |
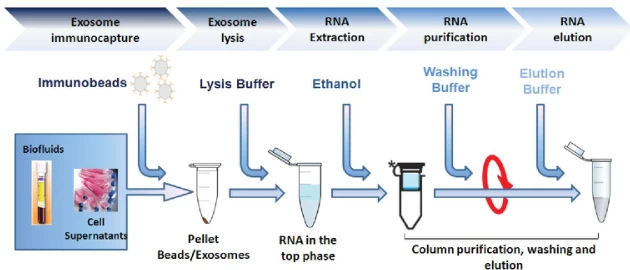
B. Key Innovations
- Simultaneous DNase treatment: gDNA removal during lysis via spin-column DNase I digestion
- Carrier RNA augmentation: MS2 bacteriophage RNA boosts low-yield samples
- Bioruptor® sonication: Ultrasonic disruption for tough tissues
III. Phase 2: RNA Binding & Contaminant Removal
A. Dominant Binding Technologies
- Silica-Membrane Adsorption (e.g., GenElute™, PureLink™)
- RNA binds to silica at high ionic strength (>4M guanidinium)
- Contaminants flow through during centrifugation
(Fig. 2: Spin-Column Mechanism)
Description: Cross-section showing RNA (blue strands) adhering to silica membrane while proteins (red) and lipids (green) pass through.
- Magnetic Bead Capture (e.g., Virus RNA Kits)
- Oligo-dT/silica-coated beads bind RNA
- Magnetic racks isolate RNA-bead complexes
- Direct-zol™ Technology
- Eliminates phenol-chloroform phase separation
- Direct TRIzol lysate application to Zymo-Spin columns
B. Critical Separation Methods
| Contaminant | Removal Technique |
|---|---|
| Genomic DNA | On-column DNase digestion |
| Proteins | Ethanol/chaotrope washes |
| Polysaccharides | PVP-40 treatment |
| Lipids | Chloroform extraction |
IV. Phase 3: Rigorous Washing Protocols
A. Standardized Wash Sequence
1. **Wash Buffer 1**: High-salt solution removes residual proteins
2. **Wash Buffer 2**: Ethanol-based (70-80%) eliminates salts
3. **DNase Wash**: Column-immobilized DNase digests DNA (optional)
Centrifugation at 12,000 rpm ensures complete contaminant removal
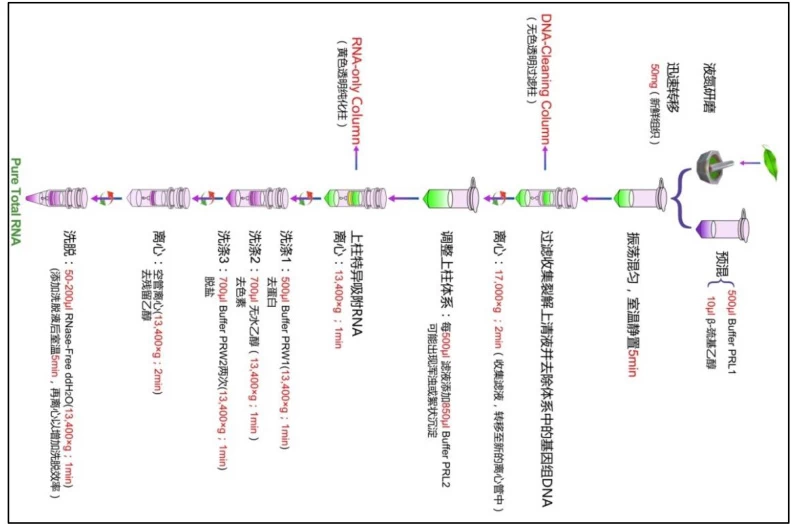
B. Specialized Washes
- FFPE samples: Extended proteinase K digestion (24h at 56°C)
- Plant materials: Double chloroform extraction for starch removal
- Viral samples: Hemoglobin inhibitor cocktails
V. Phase 4: Elution & Quality Control
A. Elution Optimization
- Low-ionic buffers: Nuclease-free water or TE buffer maximizes solubility
- Temperature enhancement: 65°C incubation improves yield
- Micro-volume calibration: 30-50µl balances concentration vs. recovery
B. Quality Verification Metrics
| Parameter | Target Value | Validation Method |
|---|---|---|
| Purity | A260/A280 ≥1.9 | Spectrophotometry |
| Integrity | RIN >7.0 | Bioanalyzer |
| DNA contamination | Ct >35 (no-RT controls) | RT-PCR |
| Yield | >1µg/mg tissue | Fluorometry |
(Fig. 3: QC Electropherogram)
Description: Bioanalyzer trace showing sharp 18S/28S rRNA peaks (RIN=8.2) vs. degraded sample (smear <RIN=5.0).
VI. Technology-Specific Workflows
A. Spin-Column Kits (e.g., PureLink™)
1. Lyse in guanidine-based buffer (5 min)
2. Load lysate onto silica column → centrifuge (1 min)
3. DNase I treatment on membrane (15 min)
4. Ethanol washes (2× centrifugations)
5. Elute in 30µl nuclease-free water (2 min)
Total time: 20 minutes
B. Magnetic Bead Kits (e.g., Virus RNA Extraction)
1. Bind RNA to oligo-dT beads (5 min)
2. Magnet separation → discard supernatant
3. Wash with 80% ethanol (2×)
4. Air-dry beads → elute with water
Throughput: 96 samples in <40 minutes
C. Phase-Free Systems (e.g., Direct-zol™)
1. Homogenize in TRIzol (3 min)
2. Directly load lysate onto column (1 min)
3. Wash/elute without phase separation
Advantage: Unbiased small RNA recovery in 7 minutes
VII. Sample-Specific Optimization
| Sample Type | Critical Adaptations | Yield Benchmark |
|---|---|---|
| Plant seeds | Buffer S1 + chloroform extraction | 5µg/100mg |
| Exosomes | Immunobead capture + specialized lysis | 15ng/mL |
| FFPE tissues | Xylene deparaffinization → extended digestion | 50% vs. fresh tissue |
| Whole blood | Leukocyte stabilization + carrier RNA | 10µg/mL |
VIII. Emerging Innovations
- CRISPR-Assisted Purification: Cas13-RNA complexes for targeted isolation

- Chemoenzymatic Methods: Solid-phase glycosylation for modified RNA
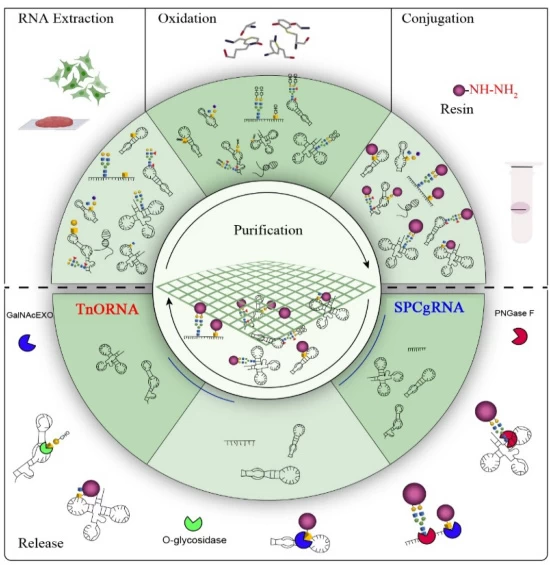
- Microfluidic Automation: Chip-based extraction (90-second processing)
- Lyophilized Field Kits: Room-temperature stable reagents
“Modern RNA extraction kits transform biological chaos into molecular precision—converting viscous cellular soups into vials of pure genetic insight.”
— Nature Biotechnology, 2025
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
