 I. Genomic Identity and Functional Dichotomy
I. Genomic Identity and Functional Dichotomy
Positive-Sense RNA Viruses (+ssRNA)
- Direct mRNA Function: The +ssRNA genome acts as immediate messenger RNA upon host cell entry. Its sequence is directly recognized by host ribosomes, enabling instantaneous protein synthesis without prior transcription .
- Infectious RNA: Purified genomic RNA can initiate infection independently (e.g., poliovirus RNA injected into host cells triggers viral replication) .
Negative-Sense RNA Viruses (-ssRNA)
- Genomic Inertness: The genome is complementary to mRNA and cannot initiate translation. Requires virion-packaged RNA-dependent RNA polymerase (RdRp) to synthesize translatable +ssRNA intermediates .
- Non-infectious RNA: Purified genomic RNA lacks infectivity due to RdRp dependency .
(Fig. 1: Genomic Translation Mechanisms)
Description: Ribosome (grey) binding directly to +ssRNA (blue) for protein synthesis. For -ssRNA (red), RdRp (yellow) first synthesizes complementary +ssRNA to enable translation.
II. Replication Strategies: Divergent Pathways
+ssRNA Replication Cycle
- Primary Translation: Genomic RNA → viral replicase (RdRp, helicases) .
- Membrane Remodeling: Forms double-membrane vesicles (DMVs) to shield double-stranded RNA (dsRNA) intermediates from host immune sensors .
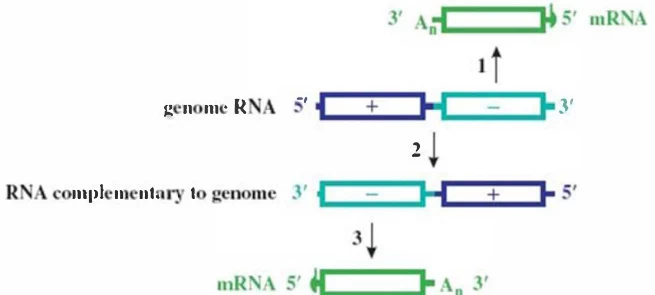
- Negative-Strand Synthesis: RdRp synthesizes complementary (-)RNA from +ssRNA template .
- Asymmetric Amplification: (-)RNA template generates 10-100× more (+)RNA progeny .
-ssRNA Replication Cycle
- Primary Transcription: Virion-carried RdRp transcribes (-)genome → monocistronic +ssRNAs .
- Cap-Snatching: Viral endonuclease “steals” 5′-methylguanosine caps from host mRNAs to prime viral transcription .
- Ribonucleoprotein (RNP) Protection: Nucleoproteins coat genomic RNA, preventing immune detection .
(Fig. 2: Replication Workflows)
Description: Top: +ssRNA replication showing DMV formation and asymmetric amplification. Bottom: -ssRNA cycle with cap-snatching and RNP assembly.
III. Structural and Evolutionary Contrasts
| Characteristic | +ssRNA Viruses | -ssRNA Viruses |
|---|---|---|
| RdRp Requirement | Synthesized de novo post-entry | Pre-packaged in virion |
| Genome Architecture | Typically non-segmented | Often segmented (e.g., influenza) |
| Mutation Rate | High (no proofreading; ~10⁻⁴ errors/base) | Lower (RNP-mediated stability) |
| Replication Site | Membrane-bound DMVs | Cytoplasmic RNP factories |
| Clinical Examples | SARS-CoV-2, Hepatitis C, Zika | Influenza, Ebola, Rabies |
(Fig. 3: Replication Complex Ultrastructure)
Description: 3D cutaway of +ssRNA DMVs (gold) with replicase complexes (purple). -ssRNA RNP complex (orange) with nucleoproteins (blue) coating genomic RNA.
IV. Diagnostic and Therapeutic Implications
A. Detection Methods
| Viral Class | Diagnostic Target | Technology |
|---|---|---|
| +ssRNA | Genomic RNA (direct detection) | RT-PCR |
| -ssRNA | Early-transcribed mRNA | NASBA/TMA amplification |
B. Antiviral Targeting
- +ssRNA Vulnerabilities:
- RdRp inhibitors: Remdesivir (chain termination)
- Protease blockers: Nirmatrelvir (inhibits polyprotein cleavage)
- -ssRNA Vulnerabilities:
- Cap-snatching inhibitors: Baloxavir (blocks influenza endonuclease)
V. Evolutionary Trade-offs
| Trait | +ssRNA Advantage | -ssRNA Advantage |
|---|---|---|
| Speed | Immediate translation (<10 min post-entry) | Controlled gene expression |
| Adaptability | High mutation rate facilitates host jumping | Segment reassortment expands host range |
| Immune Evasion | DMVs hide dsRNA from sensors | RNP complexes mask pathogen signatures |
VI. Vaccine Development Platforms
- +ssRNA Applications: Self-amplifying mRNA vaccines (e.g., COVID-19 vaccines using alphavirus replicons) .
- -ssRNA Engineering: RNP delivery for gene editing .
(Fig. 4: Vaccine Design Strategies)
Description: Left: saRNA vaccine with replicase genes (red) amplifying antigen expression. Right: RNP complex (orange) delivering CRISPR components.
“Genomic polarity dictates viral life history: +ssRNA prioritizes explosive adaptability, while -ssRNA evolves through genomic stability via structural innovation.”
— Nature Reviews Microbiology, 2025
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
