 I. Genomic Constraints and Primer Requirements
I. Genomic Constraints and Primer Requirements
Negative-sense RNA viruses (-ssRNA) possess genomes that are functionally inert upon host cell entry:
- Non-translatable Sequence: The genome is complementary to mRNA, preventing direct ribosome binding
- RdRp Dependency: Virion-packaged RNA-dependent RNA polymerase (RdRp) is essential for primary transcription
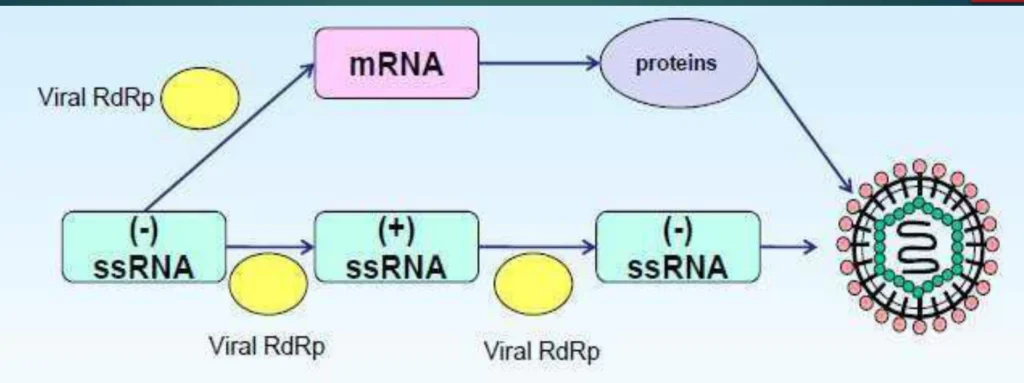
- Structural Protection: Genomic RNA is coated by nucleoprotein (NP) forming ribonucleoprotein (RNP) complexes, preventing immune recognition
(Fig. 1: Virion Entry and RNP Release)
Description: Viral envelope fusion (left) releasing RNP complexes (NP-coated RNA in purple) into cytoplasm. RdRp (yellow) bound to RNP complex.
II. Transcription Initiation: A Three-Step Mechanism
Step 1: Promoter Binding
RdRp recognizes conserved leader sequences at the 3′-end of the genome:
- Cis-acting elements: PAN (promoter-associated nucleotide) motifs in influenza viruses
- Conformational activation: RdRp unwinds RNA-NP interactions to access template
Step 2: Cap-Snatching
Unique to -ssRNA viruses, RdRp “steals” host mRNA caps:
- Endonuclease activity: Cleaves 5′-cap structures (m⁷GpppN) from host mRNAs
- Cap Grafting: Transfers caps to nascent viral transcripts via phosphodiester bonds
(Fig. 2: Cap-Snatching Mechanism)
Description: Host mRNA (blue) with m⁷G cap (gold). Viral endonuclease (red) cleaving cap. RdRp (yellow) attaching cap to viral transcript (purple).
Step 3: Processive Transcription
RdRp synthesizes monocistronic mRNAs:
- Sequential gene expression: RdRp initiates at each gene-start (GS) signal
- Attenuation at gene-end (GE) signals: Polyadenylation via RdRp “stuttering” on uridine tracts (UUUUU)
III. Transcriptional Regulation: Stop-Restart Dynamics
| Signal Type | Sequence Motif | RdRp Action |
|---|---|---|
| Gene Start (GS) | UCCU/ACNU (viral-specific) | Initiates mRNA synthesis with capped 5′-end |
| Gene End (GE) | PolyU tract | Stutters to add poly(A) tail (50-200 adenines) |
| Intergenic Region | Non-transcribed spacer | Detaches/reassembles for next gene transcription |
Critical Features:
- Polycistronic avoidance: Prevents translation of multi-gene mRNAs
- Transcriptional gradient: Proximal genes (NP, L) express >100x higher than distal genes
(Fig. 3: Stop-Restart Transcription)
Description: RdRp (yellow) synthesizing mRNA-1 (green) until GE signal (red). Reinitiation at GS signal of mRNA-2 (blue) downstream.
IV. Viral Family-Specific Variations
A. Cytoplasmic Transcription
- Rhabdoviridae (Rabies): Single RdRp entry site with sequential GS-GE processing
- Filoviridae (Ebola): Transcriptional “sliding” across overlapping genes
B. Nuclear Transcription
- Orthomyxoviridae (Influenza): Uses host nuclear RNA polymerase II for cap-snatching
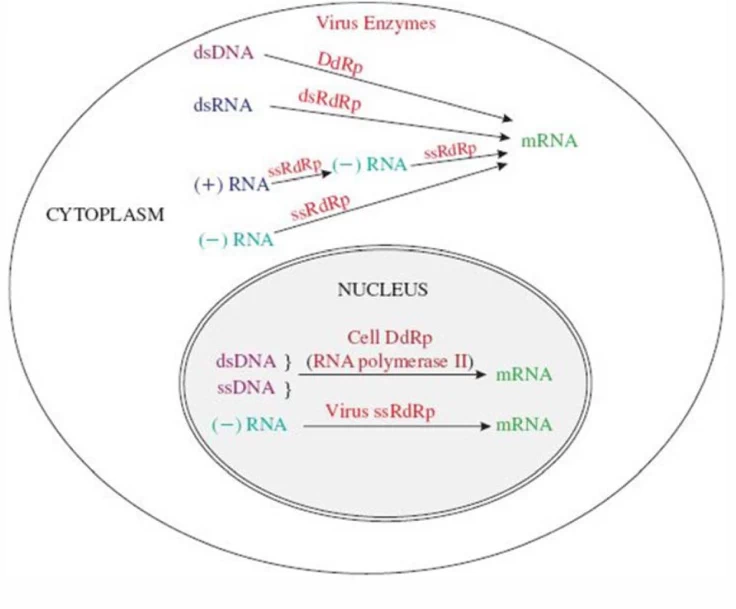
- Arenaviridae (Lassa): Ambisense coding requires both genomic and antigenomic transcription
V. Clinical Implications of Transcriptional Control
A. Antiviral Targeting
- Cap-snatching inhibitors: Baloxavir blocks influenza endonuclease
- Polymerase inhibitors: Favipiravir induces lethal mutagenesis in RdRp
B. Vaccine Design
- Attenuation strategies: GE signal mutations reduce virulence (e.g., RSV vaccine candidates)
- mRNA vaccines: Synthetic -ssRNA transcripts mimicking viral mRNAs
VI. Unresolved Mechanistic Questions
- How do RdRp complexes maintain processivity across >10,000 nt genomes?
- What triggers the switch from transcription to replication?
- Why do segmented viruses (Orthomyxoviridae) exhibit nuclear dependency?
“Negative-strand RNA viruses exemplify evolutionary ingenuity: converting genomically inert RNA into therapeutic targets through hijacked transcriptional machinery.”
— Nature Microbiology, 2025
Data sourced from publicly available references. For collaboration inquiries, contact: chuanchuan810@gmail.com.
