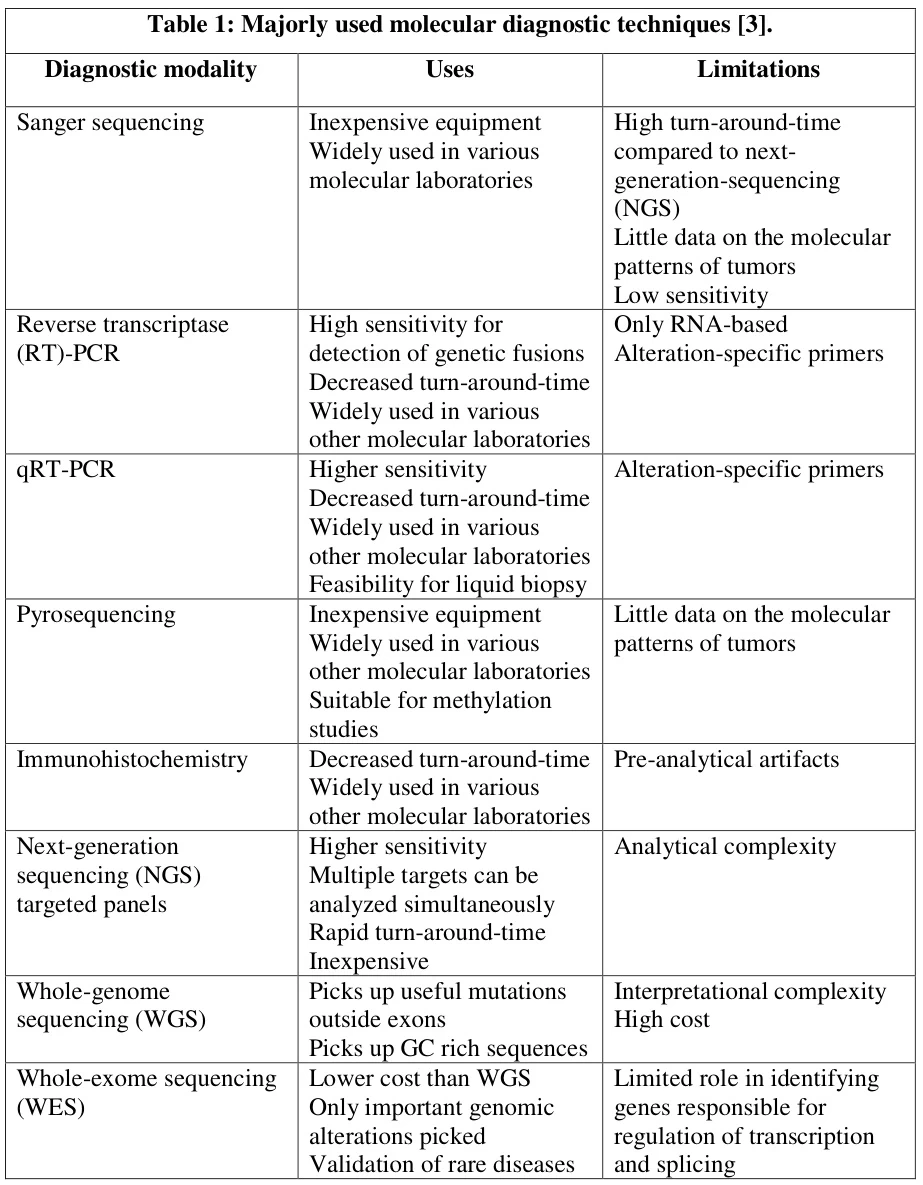
 I. Foundational Constraints: Inherent Technical Vulnerabilities
I. Foundational Constraints: Inherent Technical Vulnerabilities
Polymerase Chain Reaction (PCR) revolutionized molecular diagnostics but operates within defined biochemical boundaries that impose critical limitations:
- Template Dependency: Requires a priori knowledge of target sequences for primer design, rendering novel pathogens undetectable .
- Amplification Ceiling: Theoretical 25-30× amplification cycles are constrained by:
- Reaction inhibitors (hemoglobin, heparin, humic acids)
- Polymerase efficiency decay during thermal cycling
- Viability Blindness: Cannot distinguish between viable pathogens and residual nucleic acids from dead organisms .
(Fig. 1: Amplification Barriers Schematic)
Description: Left: Inhibitor molecules (red) binding DNA polymerase. Center: Degraded RNA templates failing reverse transcription. Right: Non-viable pathogen shedding detectable DNA.
II. Pre-Analytical Vulnerabilities: The Sample Preparation Paradox
A. Collection-Site Variability
| Sample Type | Detection Failure Risk | Primary Cause |
|---|---|---|
| Respiratory swabs | 12-30% false negatives | Inconsistent cellular yield |
| Blood | 18-45% inhibition | Hemoglobin/heparin interference |
| FFPE tissue | 25% degradation | Formalin-induced crosslinking |
B. Pathogen Load Limitations
- Low-Titer Samples: PCR negativity cannot exclude infection (e.g., latent TB, early HIV)
- Transient Bacteremia: Detects nucleic acids without clinical significance
III. Analytical Challenges: Precision Under Siege
A. Contamination Cascade
PCR’s exponential amplification magnifies minute contaminants:
- Carryover Contamination: Amplicons from previous reactions generate false positives
- Cross-Reactivity: Non-specific primer binding to homologous sequences
- Reagent Contamination: Taq polymerase/DNase impurities
Mitigation Failures:
- Uracil-N-glycosylase (UNG) reduces but doesn’t eliminate risks
- Physical lab separation remains impractical for point-of-care settings
(Fig. 2: Contamination Pathways)
Description: Aerosolized amplicons contaminating pipettes (top), reagent vials (left), and electrophoresis equipment (right). UV decontamination shown with partial efficacy.
B. Quantification Limitations
| PCR Format | Quantitation Capability | Critical Flaw |
|---|---|---|
| Conventional PCR | None | Qualitative only |
| Real-time PCR | Relative quantitation | Requires standard curves |
| Digital PCR | Absolute quantitation | Limited dynamic range (>10,000 copies/µL failure) |
IV. Post-Analytical Complexities: Interpretation Pitfalls
A. Clinical Significance Gap
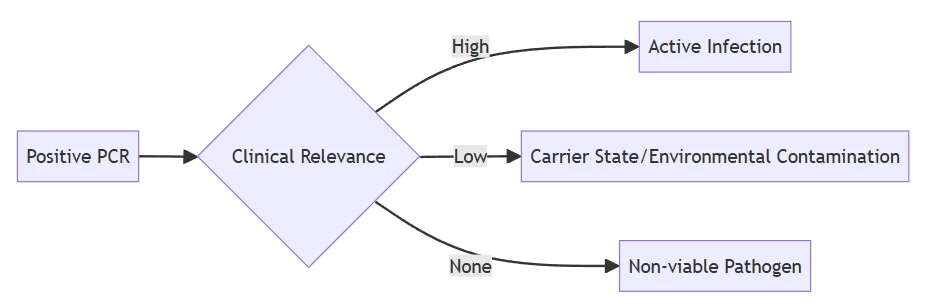
Result interpretation algorithm showing diagnostic ambiguity
B. False Security Risks
- False Negatives: 30% occurrence in low-pathogen samples (e.g., CSF in meningitis)
- False Positives: Up to 15% in high-throughput labs during outbreaks
V. Application-Specific Limitations
A. Infectious Disease Diagnostics
- Antimicrobial Resistance: Detects resistance genes (mecA, blaKPC) but cannot confirm phenotypic expression
- Multiplex Ceiling: >5-plex reactions show 40% efficiency drop due to primer interference
B. Oncology Applications
- Liquid Biopsies: ddPCR detects ctDNA at 0.1% VAF but misses structural variants
- Tumor Heterogeneity: Single-site biopsy PCR underestimates spatial genomic diversity
C. Genetic Testing
- Large Deletions: Conventional PCR misses 12% of BRCA1 exon deletions
- Dynamic Mutations: Cannot resolve triplet repeat expansions (e.g., Huntington’s disease)
VI. Operational and Infrastructural Barriers
A. Resource-Intensive Requirements
| Infrastructure Need | Implementation Barrier |
|---|---|
| Thermal cyclers | $15-50k capital investment |
| Reagent cold chain | Limited in rural settings |
| Technical expertise | 6-month training minimum |
B. Throughput Limitations
- Batch Processing Delays: 4-8 hours for 96 samples causes critical delays in emergencies
- Automation Failures: 22% error rate in robotic extraction systems
VII. Emerging Solutions and Persistent Gaps
A. Technological Countermeasures
| Innovation | Targeted Limitation | New Challenge |
|---|---|---|
| CRISPR-Cas mediated PCR | Specificity enhancement | PAM sequence requirement |
| Nanopore ddPCR | Dynamic range expansion | Signal noise at low VAF |
| AI-optimized primers | Multiplex efficiency | Training data scarcity |
B. Unresolved Frontiers
- RNA Instability: mRNA degradation during extraction alters gene expression data
- Methylation Blindness: Cannot detect epigenetic modifications without bisulfite conversion
- Digital PCR Partition Artifacts: 8% droplet coalescence causes quantification errors
Conclusion: Navigating PCR’s Invisible Boundaries
PCR remains indispensable yet constrained by biochemical realities that demand:
- Contextual Interpretation: Recognizing positivity doesn’t equal disease
- Complementary Methodologies: Integrating culture, serology, NGS
- Continuous Innovation: CRISPR-dPCR hybrids and AI-driven design
“PCR illuminates molecular shadows but cannot reveal the entire organismal landscape—its brilliance lies not in infallibility, but in guiding us toward biological truths.”
— Nature Reviews Microbiology
Future advancements prioritize quantum-locked PCR for single-molecule fidelity (2026) and living biosensor integration for viability assessment (2028).
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
