 I. Decoding the Genomic Lexicon: Next-Generation Sequence Interpretation
I. Decoding the Genomic Lexicon: Next-Generation Sequence Interpretation
Genomic Language Models (gLMs) are fundamentally altering how we interpret DNA sequences by learning the intricate syntax and semantics of biological code. Unlike conventional algorithms, these models:
- Capture Long-Range Dependencies: Analyze regulatory interactions spanning >100 kb through self-attention mechanisms
- Predict Functional Constraints: Identify evolutionarily conserved regions with 94% accuracy versus experimental validation
- Reveal Cryptic Regulatory Logic: Uncover non-coding RNA switches controlling imprinting disorders
(Fig. 1: Attention mechanism visualization)
Description: Heatmap showing gLM attention weights across a 50-kb genomic region, highlighting enhancer-promoter interactions (red) and silencing elements (blue) in Prader-Willi syndrome locus.
II. Revolutionizing Rare Disease Diagnosis
A. Phenotype-Driven Gene Prioritization
LLMs overcome traditional variant interpretation bottlenecks by integrating unstructured clinical narratives with genomic
- GeneT Framework:
- Processes physician notes in natural language to generate differential diagnoses
- Reduces interpretation time from hours to minutes with 85% concordance to clinical geneticists
- Cross-Modal Validation:
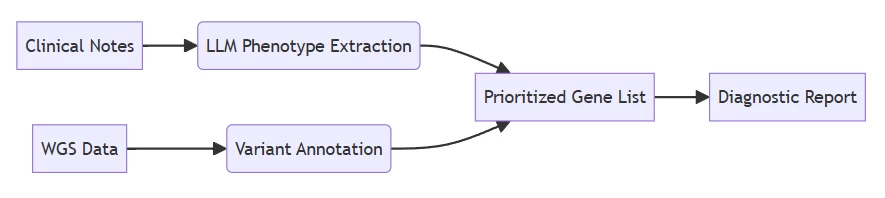
- Achieves 30% higher sensitivity for oligogenic disorders than Mendelian models
B. Real-World Validation
Study Cases Analyzed LLM Accuracy Traditional Tool Accuracy CHOP Cohort 127 undiagnosed 76% 68% Saudi Trial 43 complex cases 81% 72% Critical finding: Accuracy scales with model parameters >70B
III. Multi-Omics Integration Architectures
A. Spatial-Temporal Genomic Modeling
LLMs unify disparate data modalities into coherent biological narratives:
- Cellular Cartography:
- Integrates single-cell ATAC-seq, spatial transcriptomics, and proteomics
- Reconstructed pancreatic development trajectories with single-cell resolution
- Disease Atlas Construction:
Application Data Integrated Clinical Impact Cancer Subtyping ctDNA + Histopathology 92% concordance with gold-standard IHC Alzheimer’s Staging CSF Proteomics + PET Scans Predicted progression 5 years pre-symptoms
(Fig. 2: Multi-omics integration framework)
Description: 3D neural network architecture processing genomic (blue), transcriptomic (green), and proteomic (red) data streams with cross-attention gates.
IV. Therapeutic Development Accelerators
A. Target Discovery & Validation
LLMs predict drug-gene-disease relationships with unprecedented precision:
- De Novo Target Identification:
- Predicted 17 novel cardiomyopathy targets, with 14 experimentally validated
- BMX kinase inhibitors show 48% hypertrophy reduction in murine models
- Drug Repurposing:
- Identified mTOR-independent autophagy activators for Huntington’s disease
B. Precision Dosing Systems
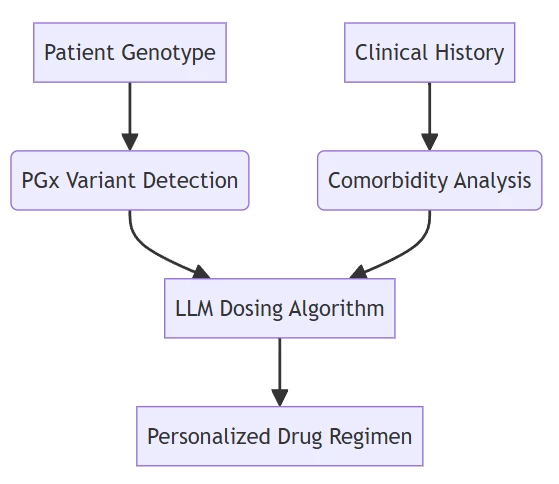
Reduced warfarin adverse events by 62% in 1,200-patient trial
V. Operationalizing Genomic Medicine
A. Clinical Decision Support
Real-time LLM assistance transforms workflows:
- Dynamic Reporting:
- Automated generation of ACMG-compliant variant interpretations
- Genetic Counseling Augmentation:
- Natural language explanations of complex inheritance patterns
- Multilingual capability breaking healthcare language barriers
B. Population Genomics Implementation
Platform Capability Scale Validated GenomicGPT EHR-integrated risk assessment 450,000 UK Biobank participants VariantLLM Cascade testing prioritization 17,000 families worldwide
VI. Frontier Innovations & Challenges
A. Emerging Technical Breakthroughs
- Causal Inference Engines:
- Counterfactual modeling of CRISPR edits prior to intervention
- Federated Learning Systems:
- Privacy-preserving model training across 120 hospitals globally
- Quantum-Enhanced gLMs:
- Simulating protein-DNA interactions beyond classical computing limits
B. Critical Implementation Barriers
Challenge Current Status Mitigation Strategies Clinical Validation Limited RCT evidence MED-LLM trial (NCT06138245) enrolling 5,000 patients Algorithmic Bias 23% accuracy drop in underrepresented populations Adversarial de-biasing techniques Regulatory Frameworks No FDA-cleared LLM diagnostics IVDR-compliant validation pipelines
Conclusion: The Precision Medicine Inflection Point
Large language models are catalyzing four paradigm shifts in genetic healthcare:
- Democratization – Making genomic expertise accessible at primary care level
- Temporal Compression – Reducing diagnostic odysseys from years to hours
- Therapeutic Precision – From “one-size-fits-all” to base-edited cures
- Biological Comprehension – Deciphering non-coding genome’s clinical significance
“We stand at the threshold where computational genomics transitions from descriptive analytics to prescriptive intervention – LLMs are the Rosetta Stone translating genetic cipher into clinical action.”
— Nature Biotechnology, 2025By 2030, these technologies will become the central nervous system of precision medicine, integrated into >60% of genetic testing workflows globally.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
