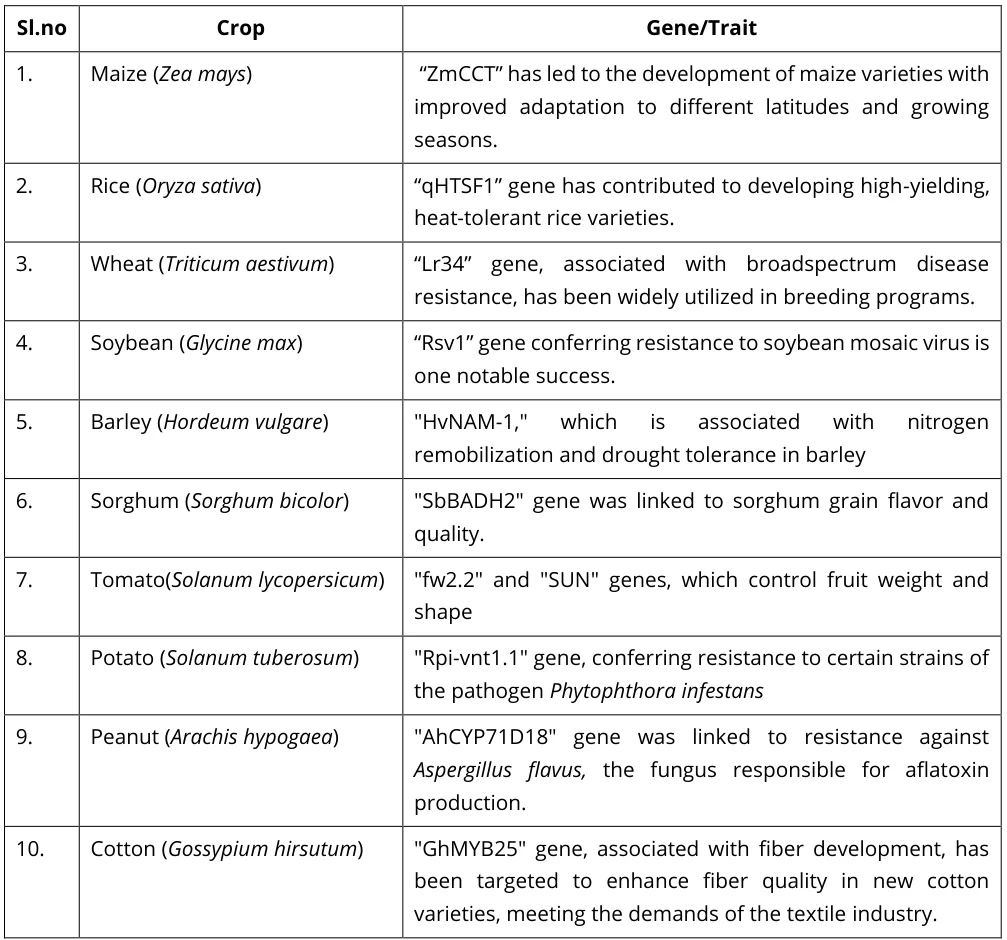
 I. Yield Architecture Genes: Engineering Productivity
I. Yield Architecture Genes: Engineering Productivity
Precision-bred alleles now optimize photosynthesis, grain filling, and reproductive efficiency:
- Ghd7 Modulators:
- Rice pleiotropic gene controlling grain number, heading date, and plant height simultaneously
- CRISPR-edited variants extend growing seasons while increasing panicle density
- NDD2-Specific Innovations:
- 1,404 novel genes identified in Glycine max cultivar “Nongdadou 2”
- GmFAD2-1A/B alleles enhance seed oil composition and stability
(Fig. 1: Molecular yield optimization)
Description: 3D protein structure of Ghd7 transcription factor (blue) bound to flowering promoter region (gold), with CRISPR-induced mutation sites (red sparks).
II. Climate Resilience Genes: Battling Environmental Stress
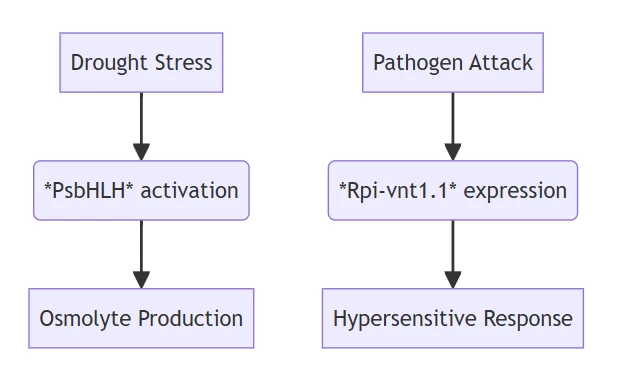
A. Abiotic Stress Networks
| Gene | Crop | Mechanism | Impact |
|---|---|---|---|
| PsbHLH | Pea | Drought-responsive TF activation | 35% yield preservation under water deficit |
| NIN/SYM | Wheat | Enhanced nitrogen fixation | 40% fertilizer reduction |
| AhCYP71D18 | Peanut | Aflatoxin resistance | Complete mycotoxin elimination |
B. Biotic Defense Systems
- CRISPR-Edited Rpi-vnt1.1:
- Confers late blight immunity in potato without fungicides
- BnITPK Alleles:
- Reduces phytic acid in rapeseed, enhancing nutritional bioavailability
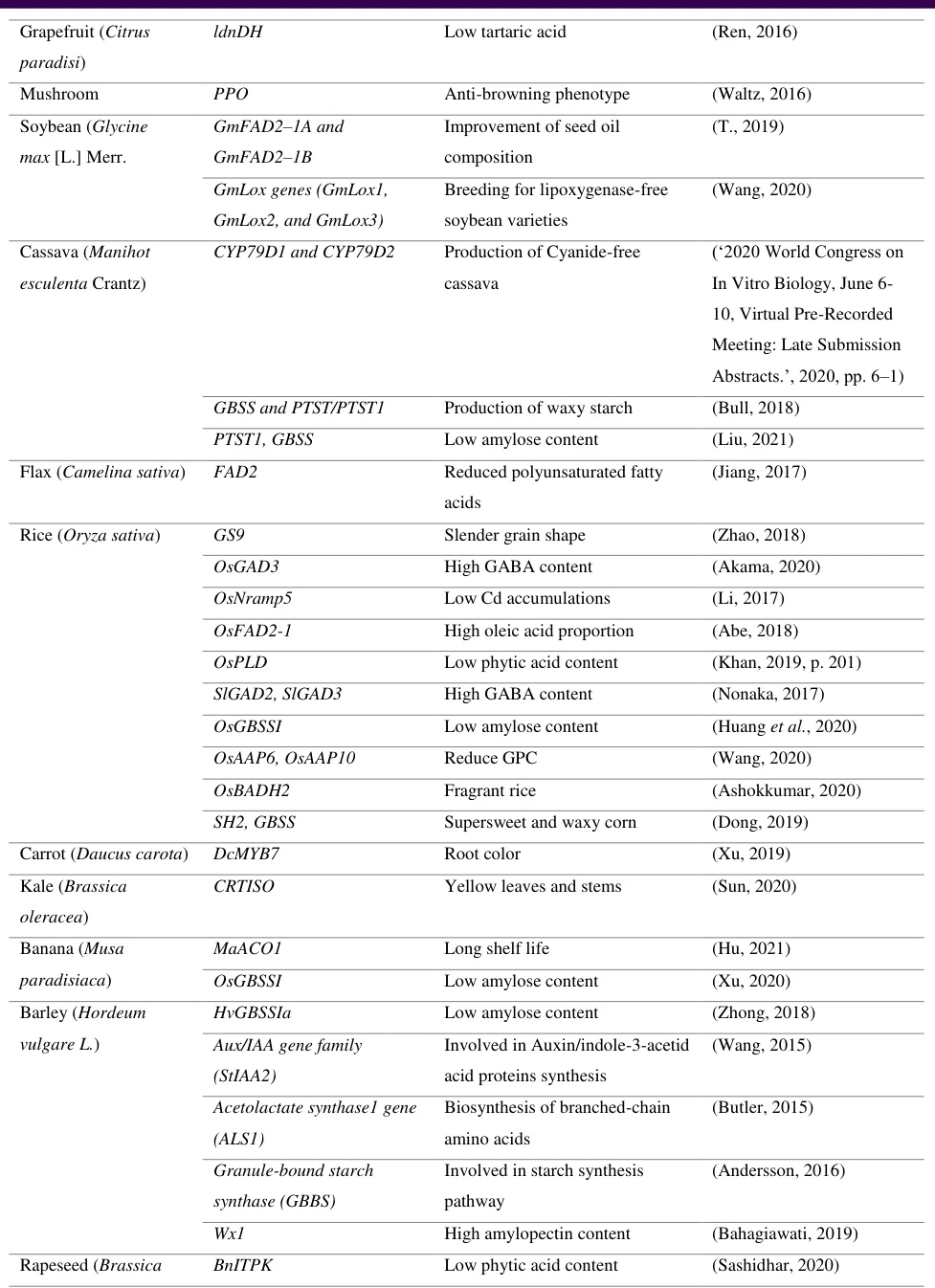
III. Nutritional Enhancement Genes: Designing Healthier Crops
A. Macronutrient Engineering
- GIF1 Variants:
- Regulates carbohydrate unloading in rice grains, increasing kernel weight by 22%
- SBEI Mutations:
- Alters starch biosynthesis in maize for high-amylose varieties (resistant starch)
B. Micronutrient Fortification
- CRTISO Editors:
- Boosts β-carotene in cabbage via carotenoid pathway engineering
- HvGBSS1a Knockouts:
- Creates low-glycemic barley for diabetic diets
(Fig. 2: Nutritional metabolic pathways)
Description: Flux balance analysis showing carbon redistribution in GIF1-edited rice vs. wild-type (red arrows indicate enhanced sucrose transport).
IV. Novel Quality Trait Genes: Beyond Basic Nutrition
A. Post-Harvest Innovations
Gene Crop Trait Market Impact MaACO1 Banana Extended shelf-life (28→45 days) 30% reduction in food waste PPO Mushroom Non-browning phenotype Enhanced fresh-cut produce sales B. Industrial Utility Genes
- GmCRY1a Orthologs:
- Soybean photoperiod adaptors enabling latitudinal expansion
- ldnDH Editors:
- Reduces tartaric acid in grapefruit for improved juice quality
V. RNA-Regulated Traits: The Epigenetic Frontier
Non-coding RNA controllers fine-tune agronomic expression:
- miR-128-3p Networks:
- Modulates nitrogen uptake efficiency in cereals via NRT regulation
- lncRNA-AA1:
- Coordinates aluminum tolerance genes in acid soils

VI. Structural Variant Hotspots: Hidden Genetic Treasures
Mega-base genomic rearrangements control complex traits:
- NDD2 Soybean SV Catalog:
- 14,237 structural variations linked to yield/quality traits
- 772 gene-body SVs directly alter protein function
- Pea Haplotype Map:
- 154.8 million SNPs guiding drought-tolerant variety development
(Fig. 3: Structural variant landscape)
Description: Circos plot showing chromosomal distribution of trait-associated SVs in soybean (red: yield traits; blue: quality traits; green: disease resistance).
VII. Commercialized Trait Systems: Market-Ready Innovations
A. Patented Gene Technologies
Trait System Developer Crop Key Gene Lepidopteran Resistance China Agricultural Univ Maize cry1Ab/cry2Ab Herbicide Tolerance DBN Maize DBN3601T GABA Enrichment EU Consortium Tomato GAD overexpression B. Pipeline Technologies (2026-2028)
- Nitrogen-Use Efficiency: OsNRT1.1B rice variants (50% N reduction)
- Photosynthetic Boosters: RCA editors accelerating carbon fixation
Conclusion: The Next-Generation Trait Universe
Agricultural genetics has evolved beyond single-gene editing to integrated trait networks:
- Precision Multi-Gene Stacks: CRISPR-facilitated pyramiding of 5+ traits
- Dynamic RNA Controllers: Tissue-specific regulation without DNA modification
- Structural Variation Mining: Exploiting natural genomic architecture
- Climate-Responsive Alleles: Real-time adaptation to changing environments
“We’ve transitioned from editing genes to engineering photosynthetic factories – where crops dynamically optimize their physiology using integrated genetic circuits responsive to environmental demands.”
— Nature Plants Editorial, March 2025The 2030 horizon anticipates quantum-biology-designed nitrogenases and AI-synthesized trait modules for zero-input agriculture.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
- NDD2 Soybean SV Catalog:
- Coordinates aluminum tolerance genes in acid soils
- GmCRY1a Orthologs:
- Creates low-glycemic barley for diabetic diets
- GIF1 Variants:
- Reduces phytic acid in rapeseed, enhancing nutritional bioavailability
