 I. Core Principles of RNA-RNA Hybridization
I. Core Principles of RNA-RNA Hybridization
RNA probes bind targets through sequence-specific Watson-Crick base pairing, forming thermodynamically stable duplexes governed by:
- Complementarity Rules
- Adenine (A) pairs with Uracil (U) via two hydrogen bonds
- Cytosine (C) pairs with Guanine (G) via three hydrogen bonds
- Single-base mismatches reduce duplex stability by 30-50%
(Fig. 1: Molecular dynamics of base pairing)
Description: Cryo-EM visualization showing H-bond formation between probe (blue) and target RNA (gold).
- Structural Compatibility
- RNA-RNA hybrids adopt A-form helices (23 Å diameter) with deep major grooves
- 2′-OH groups stabilize duplex geometry through hydrogen bonding

II. Probe Design for Target Recognition
A. Sequence Engineering Strategies
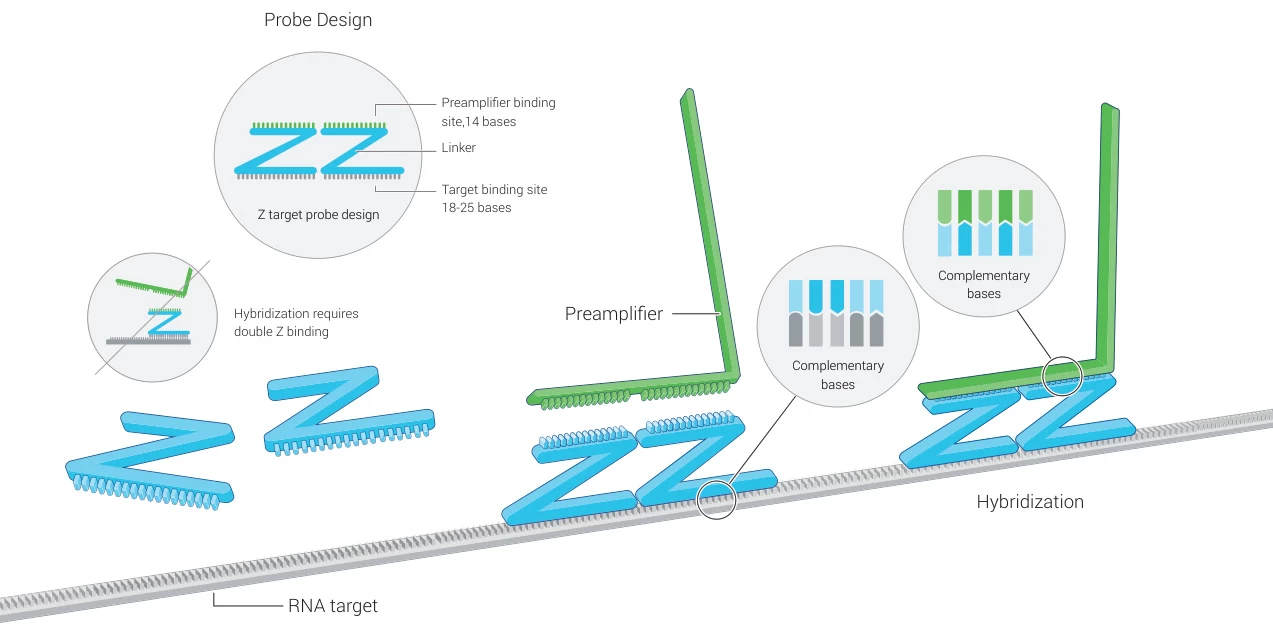
Parameter Optimal Specification Functional Impact Length 18-25 nucleotides Balances specificity and off-target binding GC Content 40-60% Precludes secondary structure formation Melting Temp (Tm) 55-70°C Ensures hybridization at physiological conditions Chemical Modifications 2′-O-methyl/LNA bases Enhances nuclease resistance (Fig. 2: Multiprobe binding architecture)
Description: RNAscope®-style “Z-probes” with target-binding regions (green), amplifier sequences (blue), and pre-amplifier sites (red).B. Thermodynamic Optimization
- Free Energy Calculations:
- ΔG ≤ -30 kcal/mol for high-affinity binding
- Penalize internal hairpins with ΔG > -5 kcal/mol
- Mismatch Discrimination:
- Single-base mismatch reduces Tm by 8-15°C
- Position mismatches near probe center for maximum specificity
III. Hybridization Dynamics & Kinetics
A. Molecular Recognition Steps
- Nucleation:
- Transient annealing of 2-4 seed nucleotides (k₁ = 10³ M⁻¹s⁻¹)
- Zippering:
- Bidirectional helix propagation (k₂ = 10⁷ M⁻¹s⁻¹)
- Branch Migration:
- Structural adjustments for optimal base stacking
(Fig. 3: Hybridization kinetics curve)
Description: Surface plasmon resonance data showing association/dissociation rates for RNA:RNA duplex formation.B. Environmental Modulators
Factor Optimal Condition Deviation Effect Temperature Tm – 20°C +5°C → 50% binding loss [Mg²⁺] 2-5 mM <1 mM → 20x slower kinetics Formamide 0-25% >40% → duplex destabilization pH 7.0-7.4 <6.0 → protonation disrupts H-bonds
IV. Validation of Binding Specificity
A. Experimental Controls
- RNase Treatment:
- Complete signal loss confirms RNA-dependent binding
- Sense/Antisense Probes:
- Antisense shows binding; sense probe serves as negative control
- Competition Assays:
- Unlabeled probes reduce signal >90% at 100x excess
B. Single-Molecule Verification
- Super-Resolution Imaging:
- dSTORM tracking of Cy5-probes confirms target colocalization
- Single-Molecule FRET:
- Real-time monitoring of hybridization dynamics
V. Advanced Recognition Systems
A. Signal Amplification Platforms
Technology Mechanism Sensitivity Gain RNAscope® Pre-amplifier → amplifier → label probe 1000x vs conventional FISH HCR Systems Hybridization chain reaction 10,000x signal amplification CRISPR-Cas13 Collateral cleavage activation Single-molecule detection (Fig. 4: HCR-based detection cascade)
Description: Target RNA initiates polymerization of fluorophore-labeled hairpins (red/green), generating amplified signal.B. Nanoscale Targeting
- Tripartite Probes:
- Folate receptor-mediated cellular delivery
- Molecular Beacons:
- Stem-loop quenching → linear activation upon binding
VI. Applications in Precision Diagnostics
A. Spatial Transcriptomics
- Tissue Section Mapping:
- Multiplexed probe panels resolve 12+ transcripts at subcellular resolution
- Single-copy viral RNA detection in clinical samples
B. Dynamic Monitoring
- Neuronal RNA Trafficking:
- Real-time tracking of β-actin mRNA in dendrites
- Viral Replication:
- RSV genome quantification during infection cycles
Conclusion: The Specificity Paradigm
RNA probes achieve molecular recognition through:
- Biophysical Precision – Watson-Crick complementarity governs target selection
- Engineered Affinity – Thermodynamic optimization enhances discrimination
- Amplified Verification – Multiprobe systems validate binding specificity
- Nanoscale Resolution – Single-molecule methods confirm true positives
“Contemporary RNA probes transcend mere detection tools – they are programmable molecular devices that interrogate RNA structure, dynamics, and localization across scales from angstroms to organisms.”
— Nature Structural & Molecular BiologyFuture innovations will focus on in vivo hybridization probes capable of blood-brain barrier penetration for neurological diagnostics.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
- Neuronal RNA Trafficking:
- Free Energy Calculations:
