 I. Core Compositional Framework
I. Core Compositional Framework
RNA probes (riboprobes) are single-stranded nucleic acid constructs engineered for target-specific binding through Watson-Crick base pairing. Their architecture integrates three fundamental components:
A. Nucleotide Backbone
- Ribose-Phosphate Chain: Forms the structural scaffold with 2′-OH groups enabling A-helix conformation
- Sequence Design: 20-50 base antisense regions complementary to target mRNA/RNA
- Modified Bases: Selenophene-uracil for X-ray diffraction or fluorophore conjugation
(Fig. 1: Nucleotide-level anatomy of an RNA probe)
Description: Molecular visualization highlighting ribose 2′-OH groups (red), phosphate linkages (yellow), and selenophene-uracil modification (green).
B. Signal Transduction Modules
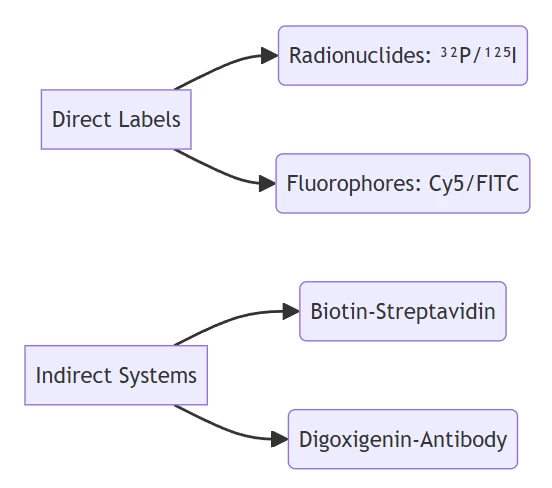
Fluorophore pairs (e.g., FAM/TAMRA) enable FRET-based detection with <2 nm distance dependency
II. Synthesis Methodologies
A. Enzymatic Production
| Method | Mechanism | Output |
|---|---|---|
| In Vitro Transcription | Phage polymerases (T7/SP6) + DNA template | Full-length ssRNA probes |
| End-Labeling | Kinase-mediated ³²P transfer to 5′-end | Radioactive probes |
| In Vivo Expression | Vector-based transcription in host cells | Cell-compatible probes |
B. Chemical Synthesis
- Phosphoramidite Chemistry: Solid-phase synthesis of oligonucleotide probes
- Hybrid Systems: Y-shaped DNA/RNA chimeras for in vivo hybridization chain reactions
(Fig. 2: IVT workflow for probe synthesis)
Description: Diagram showing plasmid linearization, RNA polymerase binding, and NTP incorporation.
III. Structural Modifications for Enhanced Functionality
A. Conformation-Sensitive Probes
- SHAPE Reagents: 2′-OH acylation to map RNA folding
- Fe(II)-EDTA Complex: Hydroxyl radical cleavage for tertiary structure analysis
B. Cellular Delivery Enhancers
| Component | Function | Example |
|---|---|---|
| Folate Conjugates | Receptor-mediated endocytosis | DNA-peptide folate probes |
| Charge-Neutral Backbones | Membrane permeability | Phosphorothioate linkages |
| Tripartite Assemblies | In vivo self-assembly | Y-probes with H1/H2 hairpins |
(Fig. 3: Tripartite Y-probe for live-cell imaging)
Description: 3D model showing folate targeting moiety (purple), RNA-binding arms (blue/green), and fluorophore (yellow).
IV. Probe-Target Interaction Dynamics
A. Hybridization Specificity
- A-Form Helix Stability: RNA-RNA duplex ΔG = -30 kcal/mol vs. DNA-DNA -22 kcal/mol
- Mismatch Discrimination: Single-base resolution via melting curve analysis
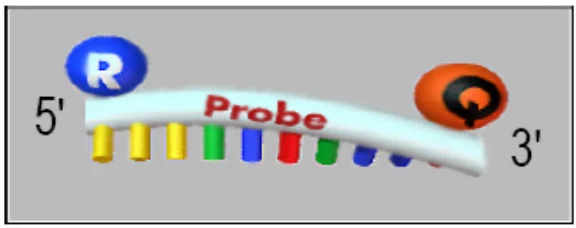
B. Turn-ON Mechanisms
- Molecular Beacons:
- Stem-loop quenched probes → Linear activation upon binding
- HCR Amplification:
- Target-triggered polymerization → 10,000x signal amplification
V. Functional Classes & Applications
A. Detection-Optimized Probes
| Type | Key Feature | Application |
|---|---|---|
| FISH Probes | Multiplexed fluorophores | Subcellular RNA mapping |
| RNase Protection | Nuclease resistance | Transcript quantification |
| Cryo-EM Probes | Heavy-atom labels | Ribosomal decoding site imaging |
B. Therapeutic Probes
- CRISPR-Cas13 Integration: Collateral cleavage activation
- Photocaged Designs: 405 nm-triggered activation in neurons
Conclusion: Convergence of Form and Function
RNA probes exemplify four structural principles:
- Modularity: Decoupled target-binding and signal domains
- Adaptability: Base modifications tailoring probe reactivity
- Bioorthogonality: Minimal interference with native RNA function
- Programmability: HCR/CAS-based signal amplification cascades
“Contemporary RNA probes transcend mere detection tools – they are programmable molecular devices that interrogate RNA structure, dynamics, and localization across scales from angstroms to organisms.”
— Nature Structural & Molecular Biology
Future innovations will focus on in vivo folded RNA probes capable of blood-brain barrier penetration for neurological disorder diagnostics.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
