 I. Core Conceptual Framework
I. Core Conceptual Framework
RNA probes (riboprobes) are single-stranded RNA molecules engineered to bind complementary nucleic acid sequences through Watson-Crick base pairing. These probes serve as molecular detection tools with transformative applications across genomics, diagnostics, and live-cell imaging. Three defining characteristics distinguish them:
- Molecular Architecture
- Synthesized via in vitro transcription from DNA templates using phage polymerases (T7, T3, SP6)
- Incorporate modified nucleotides (biotin, digoxigenin, fluorophores) for signal generation
(Fig. 1: Structural anatomy of a typical RNA probe)
Description: Molecular visualization showing fluorophore-conjugated ribonucleotides with complementary target-binding region highlighted.
- Hybridization Superiority
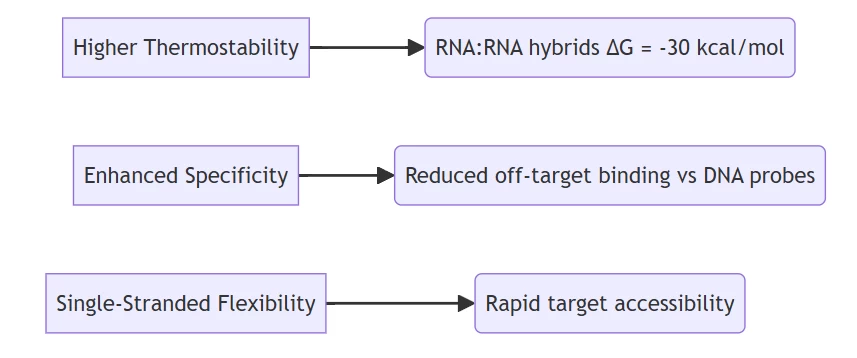
- RNA:RNA duplexes exhibit 35% greater stability than DNA:DNA equivalents due to A-form helix geometry
II. Technical Specifications & Synthesis
A. Production Workflow
Stage Critical Components Quality Control Template Design Phage promoter + target sequence BLAST specificity verification In Vitro Transcription NTP mix + modified nucleotides HPLC purity >95% Purification DNase treatment + ethanol precipitation A260/A280 = 1.9-2.1 Validation Target-spiked negative controls Signal:noise >10:1 (Fig. 2: RNA probe synthesis pipeline)
Description: Diagram illustrating sequential stages from plasmid linearization to purified probe preparation.B. Modification Technologies
- Direct Labeling:
- 32P/125I isotopes for autoradiography (sensitivity to 0.01 attomoles)
- Fluorescein/Cy5 for real-time imaging
- Indirect Detection:
- Biotin-streptavidin amplification (1000x signal enhancement)
- Digoxigenin-antibody conjugates
III. Functional Applications
A. Diagnostic & Research Methodologies
Technique Probe Function Detection Limit Northern Blotting Target mRNA quantification 0.1 pg RNA In Situ Hybridization (ISH) Spatial gene mapping Single-copy resolution RNase Protection Transcript structure analysis 5-fold > DNA probes Microarray Screening Pathogen signature detection 50 pathogens/mL B. Live-Cell Dynamics Monitoring
- Neuroscience Applications:
- Dendritic GAP43 tracking using Cy3-riboprobes (
- Real-time neurotransmitter response imaging
- Viral Replication Studies:
- RSV genome trafficking in respiratory epithelia
(Fig. 3: Multiplexed RNA probes in neuronal tissue)
Description: Confocal micrograph showing simultaneous detection of β-actin (green) and tau mRNA (red) in human hippocampus.
- RSV genome trafficking in respiratory epithelia
IV. Comparative Advantage Matrix
Parameter RNA Probes DNA Probes cDNA Probes Hybridization Affinity ★★★★★ ★★★☆☆ ★★★★☆ Thermal Stability 85°C melting temp 70°C melting temp 75°C melting temp Synthesis Cost $25/reaction $8/reaction $40/reaction Nuclease Resistance Requires RNase inhibitors High Moderate Single-Cell Resolution Compatible with super-resolution imaging Limited by background Limited by penetration
V. Emerging Innovations
- Smart Probes
- CRISPR-Cas13a-integrated systems for amplified detection
- FRET-based molecular beacons with <5 nm resolution
- Clinical Translation
- NSCLC circulating tumor RNA signatures (90% specificity)
- Rapid Zika/HIV co-infection screening chips
- Synthetic Biology
- RNA origami scaffolds for multiplexed pathogen capture
- Photocaged probes controlled by 405 nm irradiation
(Fig. 4: CRISPR-RNA probe fusion system)
Description: Molecular schematic showing Cas13a collateral cleavage activating fluorescence upon target binding.
Conclusion: The Evolving Paradigm
RNA probes represent the convergence of three transformative capabilities:
- Molecular Precision – Single-base mismatch discrimination
- Dynamic Monitoring – Real-time transcriptional imaging
- Clinical Scalability – Point-of-care diagnostic integration
“Riboprobes transform nucleic acid detection from bulk biochemical assays into a spatially resolved molecular cartography – mapping genetic activity across tissues, cells, and subcellular compartments with nanometer precision.”
— Nature Reviews Molecular BiologyOngoing advances aim to develop in vivo RNA nanoprobes capable of crossing the blood-brain barrier for neurological disorder diagnostics by 2027.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
