 I. Foundational Principles of RNA Contamination Control
I. Foundational Principles of RNA Contamination Control
RNase decontamination is paramount due to the enzyme’s ubiquitous presence and extreme stability. Key biochemical mechanisms include:
- RNase Inactivation Chemistry
- β-mercaptoethanol (0.1-1%) disrupts disulfide bonds in RNases, irreversibly denaturing their catalytic sites
- Guanidinium thiocyanate (>4M concentration) denatures proteins while maintaining RNA integrity
-
- (Fig. 1: Molecular mechanism of RNase inactivation)
Description: 3D visualization showing β-mercaptoethanol disrupting RNase tertiary structure through disulfide bond cleavage.
- (Fig. 1: Molecular mechanism of RNase inactivation)
- Environmental Control Hierarchy
- Establish physical isolation zones with positive air pressure
II. Pre-Extraction Phase: Proactive Contamination Prevention
A. Workspace & Equipment Preparation
Surface/Item Decontamination Protocol Scientific Rationale Benchtops RNaseZap® treatment + UV irradiation (30 min) Degrades RNases via alkaline hydrolysis Glassware 180°C baking for 4 hours Thermal denaturation of RNases Plasticware 0.1% DEPC-treated water immersion (2 hr) Carbethoxylation of histidine residues Centrifuges Pre-cool to 4°C; seal rotor buckets Prevents aerosol contamination B. Sample Handling Protocols
- Biological Specimens:
- Snap-freeze tissues in liquid N₂ within 30 seconds of collection
- Preserve liquid samples in RNAlater™/RNAstable® for room-temperature transport
- Personal Protective Equipment:
- Double-glove with frequent changes (every 15 min)
- Wear surgical masks to prevent salivary RNase contamination
(Fig. 2: Contamination vector analysis in RNA workflows)
Description: Infographic ranking contamination sources: skin (42%), aerosols (33%), equipment (25%)
III. Extraction Phase: Critical Control Points
A. Organic Phase Separation Management
Contaminant Type Detection Sign Corrective Protocol Genomic DNA A260/A280 >2.0 On-column DNase I digestion (37°C/15 min) Phenol residuals A260/A230 <1.8 25% increased ethanol wash volume Protein carryover Precipitated pellet cloudiness Repeat acid-phenol extraction B. Phase-Lock Techniques
- Aqueous Phase Recovery:
- Leave 2mm clearance above interphase during pipetting
- Use phase-lock gel tubes for absolute separation
- Silica Column Optimization:
- Centrifuge at ≤8,000g to prevent membrane rupture
- Validate binding capacity before processing large samples
IV. Post-Extraction Quality Assurance
A. Integrity & Purity Validation
Assessment Method Acceptance Criteria Contamination Indicators Bioanalyzer RIN ≥8.0; 28S:18S=2:1 RIN ≤6.0; DV200<30% Spectrophotometry A260/A280=1.8-2.0 Protein/organic deviations PCR amplification Ct ≤30 for housekeeping genes DNA contamination 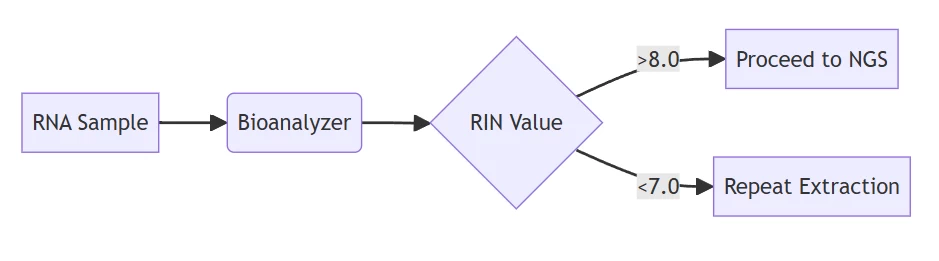
B. Storage & Stability Protocols
- Short-term: -80°C with RNase inhibitors (RNAsin®) in single-use aliquots
- Long-term: Anhydrobiotic stabilization at room temperature
V. Specialized Sample Contamination Control
A. Challenging Matrices
Sample Type Unique Contaminants Targeted Solutions Plant tissues Polysaccharides/polyphenols CTAB buffer + 2% PVP-40 FFPE samples Formaldehyde crosslinks Extended Proteinase K (24h/56°C) Whole blood Hemoglobin/heme Leukocyte separation filters B. Low-Input Applications
- Microfluidic Isolation:
- Integrated lysis-to-elution in sealed chips
- Picoliter-scale chambers eliminate cross-contamination
(Fig. 3: Microfluidic RNA extraction chip)
Description: Sealed chip architecture with separate lysis (red), binding (blue), and elution (green) chambers.
VI. Advanced System Solutions
A. Automated Platforms
- Robotic Liquid Handlers:
- Enclosed systems with HEPA filtration
- UV decontamination between samples
- Closed-Cartridge Systems:
- Pre-packaged reagent kits with integrated waste
B. Next-Gen Stabilizers
- RNAstable®:
- Water-soluble barriers create anhydrobiotic environment
- Cryo-protective Additives:
- Trehalose maintains RNA integrity during freeze-thaw
Conclusion: The Zero-Contamination Framework
Achieving RNase-free RNA requires:
- Preemptive Neutralization:
- Surface decontamination + chemical inhibitors
- Physical Barriers:
- Dedicated workspaces + sealed systems
- Process Vigilance:
- Phase-lock separation + capacity validation
- Post-Isolation Verification:
- RIN/DV200 thresholds + aliquot storage
“RNA extraction contamination control isn’t a protocol – it’s a holistic discipline requiring relentless attention to molecular enemies at every interface between sample and solution.”
— Journal of Molecular DiagnosticsFuture innovations will focus on AI-driven contamination monitoring systems with real-time degradation alerts.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
