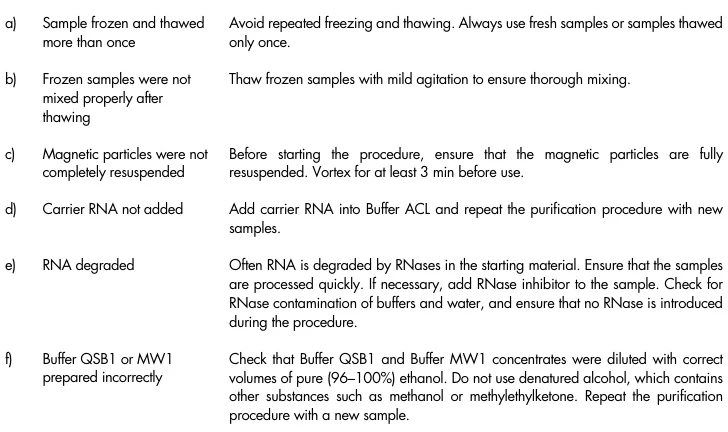
 I. Pre-Extraction Phase: Sample Integrity Preservation
I. Pre-Extraction Phase: Sample Integrity Preservation
RNA degradation begins immediately post-collection, necessitating rigorous stabilization protocols:
- RNase Inactivation Strategies
- Treat surfaces with RNase-specific decontaminants (e.g., RNaseZap®) and use certified RNase-free consumables
- Add β-mercaptoethanol (0.1-1%) or DTT to lysis buffers to denature RNases

-
-
- (Fig. 1: Molecular mechanism of RNase inhibition)
Description: 3D visualization showing β-mercaptoethanol disrupting RNase disulfide bonds.
- (Fig. 1: Molecular mechanism of RNase inhibition)
- Sample Handling Protocols
Sample Type Optimal Protocol Critical Modifications Tissues Snap-freeze in liquid N₂ within 30 sec Avoid repeated freeze-thaw cycles Blood PAXgene/RNAstable® tubes Prevent hemolysis Cultured Cells Complete media removal before lysis Residual FBS contains RNases

graph TD
A[Fresh Sample] –> B{Stabilization Method}
B –>|Tissues| C[Liquid N₂ Flash-Freeze]
B –>|Liquid Samples| D[Chemical Stabilizers]
C –> E[-80°C Storage]
D –> F[Room Temp Preservation] -
- —
### **II. Extraction Phase: Yield & Purity Optimization**
#### **A. Low RNA Yield Solutions**
| **Cause** | **Detection Method** | **Corrective Action** |
|———–|———————-|———————-|
| Insufficient lysis | Visible tissue pellets | Increase mechanical disruption: <br>- Cryogenic grinding with liquid N₂ <span data-key=”6″ class=”reference-num” data-pages=”undefined”>12</span> <br>- Bead-beating ≥90 sec <span data-key=”7″ class=”reference-num” data-pages=”undefined”>9</span> |
| Excessive starting material | Gel electrophoresis smear | Reduce tissue input by 30-50% <span data-key=”8″ class=”reference-num” data-pages=”undefined”>1</span><span data-key=”9″ class=”reference-num” data-pages=”undefined”>6</span> |
| Incomplete elution | Low A260 readings | Add 30-50µl RNase-free H₂O to membrane <br>Incubate 10 min before centrifugation <span data-key=”10″ class=”reference-num” data-pages=”undefined”>9</span> |
*(Fig. 2: Silica membrane saturation dynamics)*


*Description: SEM micrograph showing optimal (left) vs. insufficient (right) RNA binding to silica matrix.*#### **B. Contamination Management**
1. **Genomic DNA Contamination**:
– Column-integrated DNase treatment (37°C/15 min) <span data-key=”11″ class=”reference-num” data-pages=”undefined”>8</span>
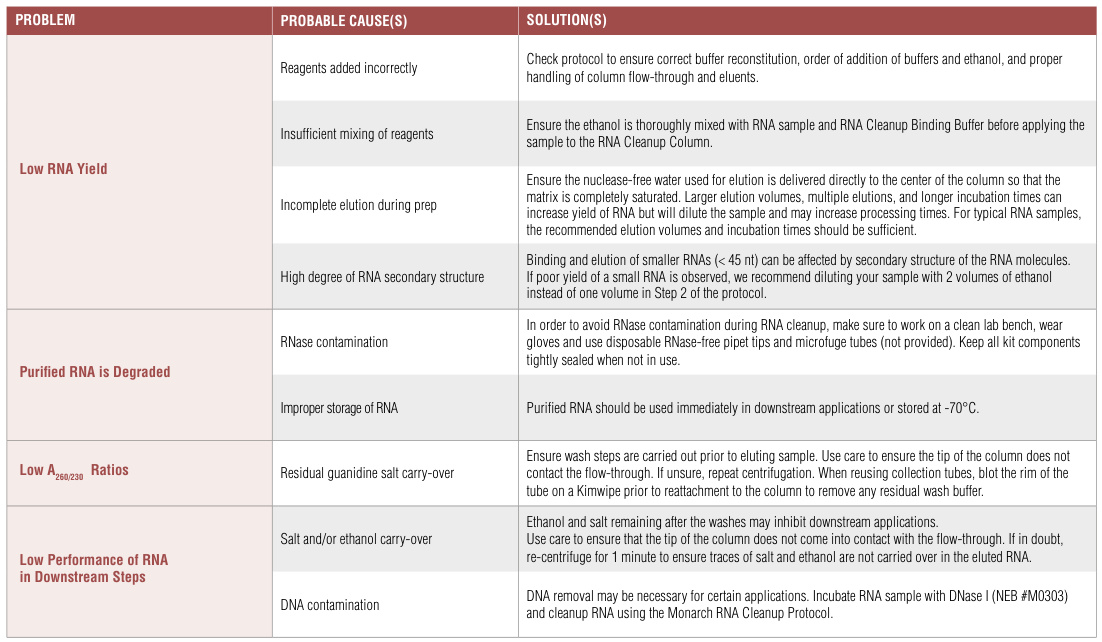
– Design intron-spanning primers for downstream applications <span data-key=”12″ class=”reference-num” data-pages=”undefined”>12</span>
2. **Organic/Particulate Contaminants**:| **A260/A230** | **Contaminant Type** | **Solution** |
|—————|———————-|————–|
| <1.8 | Polysaccharides | CTAB buffer for plant tissues <span data-key=”13″ class=”reference-num” data-pages=”undefined”>14</span> |
| <2.0 | Guanidinium salts | Additional ethanol washes <span data-key=”14″ class=”reference-num” data-pages=”undefined”>8</span> |—
### **III. Post-Extraction Phase: Integrity & Stability**
#### **A. RNA Degradation Prevention**
– **Electrophoretic Validation Standards**:| **Sample Type** | **Intact RNA Indicator** | **Degradation Threshold** |
|—————-|————————–|—————————|
| Mammalian cells | 28S:18S = 2:1 | Ratio <1.0 <span data-key=”15″ class=”reference-num” data-pages=”undefined”>7</span> |
| FFPE tissues | DV200 >30% | DV200 <20% <span data-key=”16″ class=”reference-num” data-pages=”undefined”>6</span> |
“`mermaid
graph LR
A[RNA Sample] –> B(Bioanalyzer)
B –> C{RIN Value}
C –>|>8.0| D[Proceed to NGS]
C –>|<7.0| E[Repeat Extraction]B. Storage & Handling Protocols
Duration Storage Conditions Preservation Additives <1 week -20°C 0.1 mM EDTA >1 month -80°C RNase inhibitors Transport Room temp RNAstable®/Anhydrobiotic tech Critical Practice: Aliquot RNA to limit freeze-thaw cycles to ≤3
IV. Specialized Sample Optimization
A. Challenging Sample Matrix Solutions
Sample Type Primary Challenge Optimized Protocol Plant tissues Polysaccharide contamination CTAB buffer + 2% PVP-40 Adipose tissue Lipid interference Double-volume chloroform washes FFPE samples Crosslinked RNA Extended Proteinase K digestion (24h/56°C) Whole blood Hemoglobin inhibition Leukocyte separation filters B. Low-Input Applications
- Carrier Enhancement:
- Add 1µg glycogen or linear acrylamide during precipitation
- Reduce elution volume to ≤30µl
- Microfluidic Platforms:
- Integrated extraction with picoliter-scale chambers
(Fig. 3: Microfluidic RNA extraction chip)
Description: Microchannel network with temperature-controlled lysis (4°C) and binding zones (22°C).
V. Quality Control Framework
A. Spectrophotometric Standards
Parameter Acceptable Range Corrective Action A260/A280 1.8-2.0 Phenol-chloroform re-extraction A260/A230 >2.0 25% increased ethanol wash volume Concentration >50 ng/µl Carrier RNA addition B. Electrophoretic Verification
- Agarose Gel Analysis:
- Denaturing gels with sharp 28S/18S ribosomal bands
- RIN/DV200 Validation:
- Require RIN>7 for RNA-seq; DV200>30% for FFPE
Conclusion: The RNA Integrity Preservation Protocol
Achieving high-quality RNA requires addressing three critical dimensions:
- Preemptive RNase Neutralization
- Chemical inhibitors (β-mercaptoethanol/DTT)
- Surface decontamination protocols
- Sample-Specific Lysis Optimization
- Mechanical disruption enhancements
- Contaminant-specific buffers (CTAB for plants)
- Post-Isolation Vigilance
- Aliquoted -80°C storage
- RIN/DV200 quality thresholds
“RNA extraction is not a protocol – it’s a chain of custody where each link must guard against the omnipresent threat of RNase degradation.”
— Molecular Systems BiologyFuture innovations will focus on integrated microfluidic systems performing extraction, QC, and library prep in <30 minutes with real-time degradation monitoring.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
- Carrier Enhancement:
