 I. Understanding Ribosome Load Dynamics
I. Understanding Ribosome Load Dynamics
mRNA ribosome load (MRL) represents the number of actively translating ribosomes per transcript at a given time, directly determining protein synthesis efficiency. Key biochemical principles include:
- Initiation-Extension Balance
- Translation initiation rate controls ribosome assembly on mRNA
- Slow elongation causes ribosomal traffic jams, increasing MRL but risking premature mRNA decay
(Fig. 1: Ribosome density vs. mRNA stability relationship)
Description: Inverse correlation curve showing high MRL accelerating degradation while moderate loading maximizes protein output.
- Structural Determinants
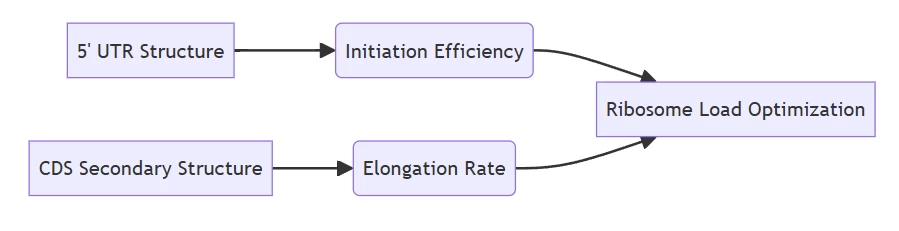
- 5′ UTR governs initiation frequency; coding sequence (CDS) folding modulates ribosomal movement
II. Sequence Engineering Strategies
A. 5′ UTR Computational Design
Approach Mechanism Efficacy UTRGAN (AI Generator) Generates UTRs with optimized Kozak contexts 32% expression increase; 12% MRL boost PERSIST-seq Screening High-throughput testing of UTR libraries Identifies stability-translation tradeoffs Free Energy Optimization Adjusts unfolding energy around start codon Enhances 43S ribosomal scanning (Fig. 2: UTRGAN’s adversarial network architecture)
Description: Generator creates UTR sequences while discriminator evaluates biological plausibility.B. CDS Optimization Techniques
- Codon Usage Modulation
- Moderate optimality: Avoid extreme codon bias to balance elongation rate
- Structured pause sites: Introduce selective secondary structures to prevent traffic jams
- Secondary Structure Engineering
- Algorithms design CDS folding with ≤-30 kcal/mol stability near start codon
III. AI-Driven Predictive Modeling
A. Deep Learning Frameworks
- Orthrus foundation models: Predict MRL from sequence with 89% accuracy using evolutionary patterns
- Ribosome flux calculators: Simulate ribosomal movement based on codon-specific elongation rates
B. Experimental Validation Integration
Technology Advantage Application RiboLace Captures actively translating ribosomes Eliminates non-productive complexes Ribo-Calibration Quantifies absolute ribosome numbers Measures initiation rates precisely Nascent Ribo-Seq Tracks ribosomal loading kinetics Reveals dynamic responses to stimuli (Fig. 3: RiboLace magnetic capture workflow)
Description: Puromycin-based selection of translating ribosomes via GFP-tagged ribosomal proteins.
IV. Biological Process Optimization
A. Ribosome Pool Management
- RsfS factor modulation: Prevents idle 70S ribosome accumulation during nutrient stress
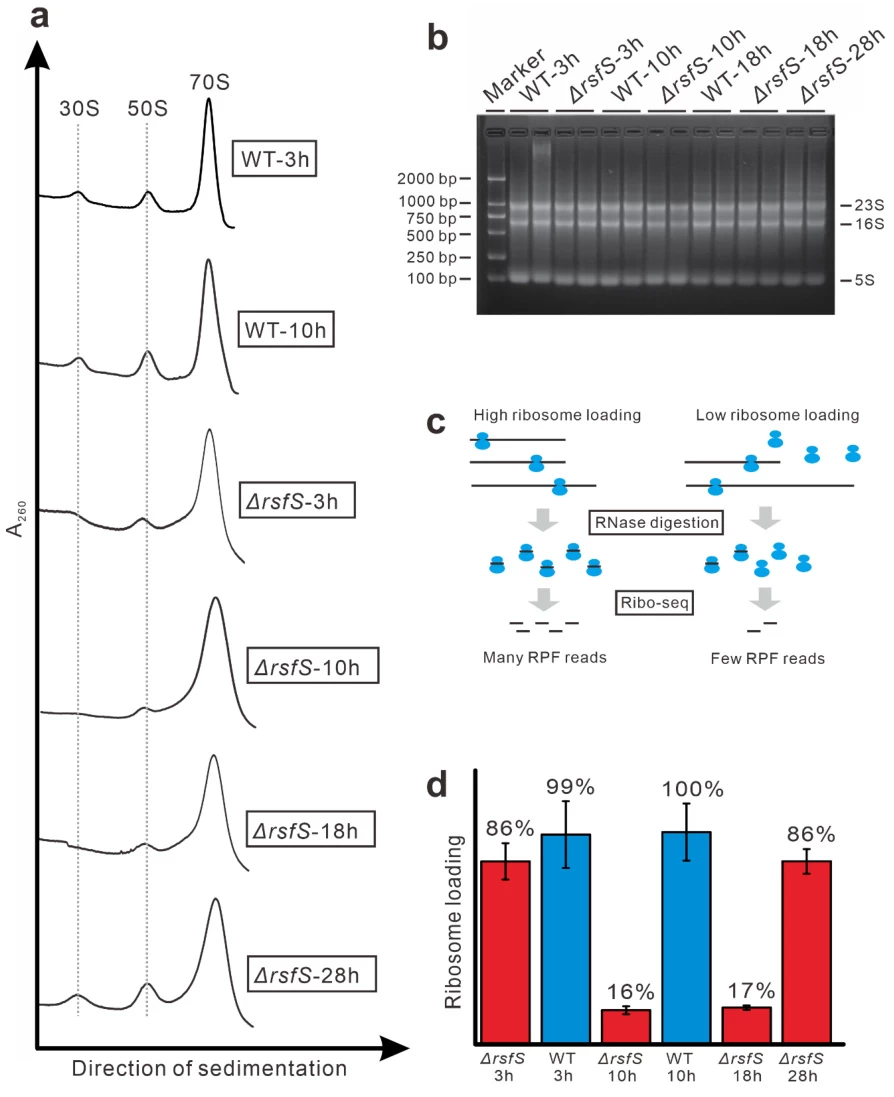
ER-bound ribosome enrichment: Upregulates during UPR to enhance secretory protein synthesis
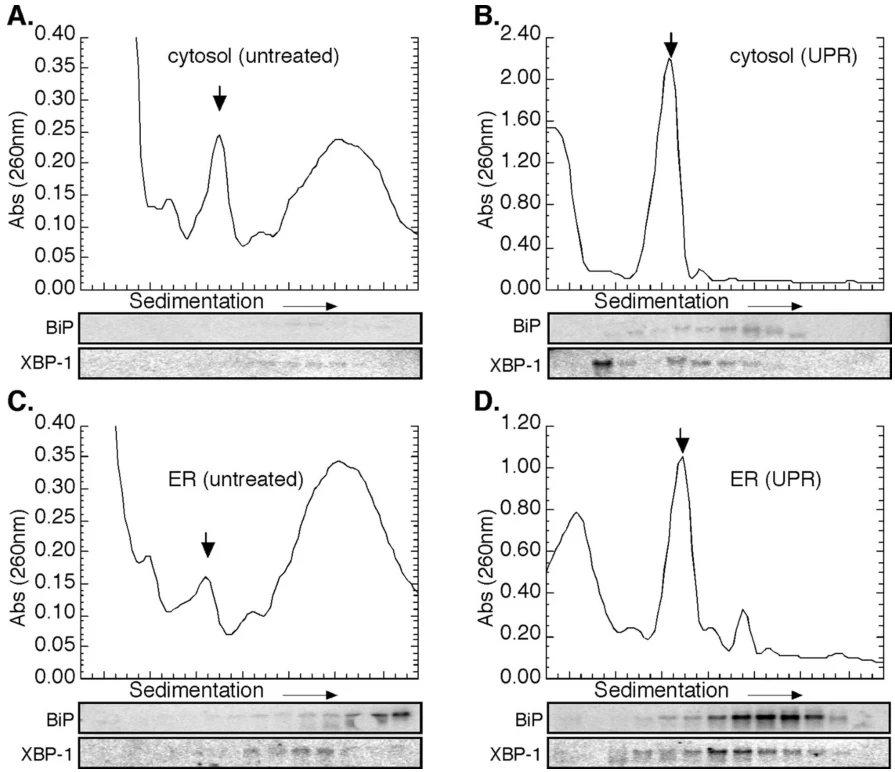
B. mRNA Stability Co-Optimization
Parameter High MRL Risk Optimized Approach mRNA half-life Accelerated decay Moderate codon optimality + UTR stabilization Decay pathways TDD/DDD activation Structure-guided avoidance of ribosome collisions
V. Integrated Workflow Implementation
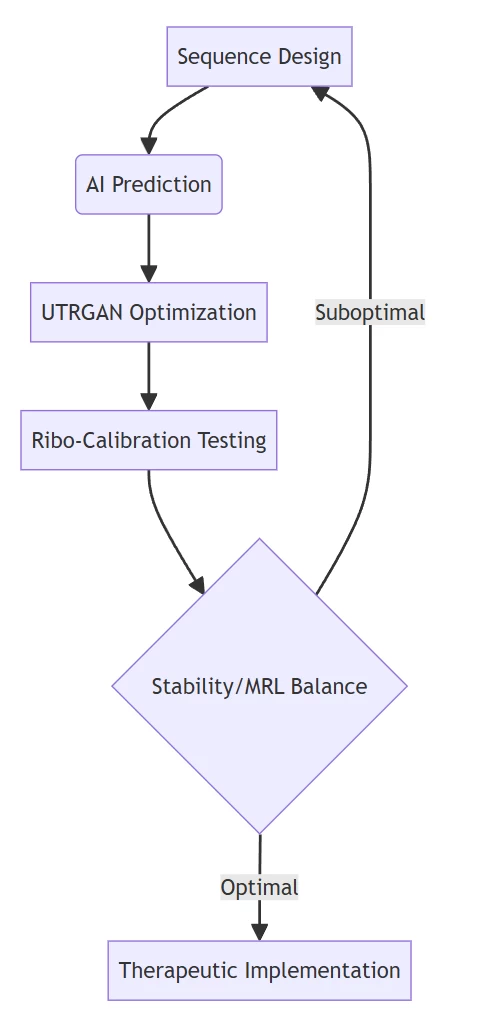
Iterative optimization loop for maximal protein output
Key Performance Metrics:
Method MRL Increase Protein Output Gain UTRGAN 12% 32% Codon Deoptimization -15% MRL +40% total protein Ribo-Calibration N/A 5-fold measurement accuracy
VI. Emerging Frontier: Dynamic Regulation
- Stress-Responsive UTRs
- Self-adjusting structures that modulate MRL during nutrient deprivation
- Riboswitch Integration
- Ligand-controlled ribosome loading for precise therapeutic dosing
- Closed-Loop mRNA Systems
- Real-time MRL monitoring via nanopore ribosome mapping
(Fig. 4: Thermo-responsive ribosome loading mechanism)
Description: mRNA thermometer structure unfolding at fever temperatures to increase antibiotic synthesis.
Conclusion: The Balanced Loading Paradigm
Maximizing protein output requires:
- AI-Powered Design: UTRGAN for initiation optimization
- Elongation Engineering: Controlled codon usage and structured pauses
- Precision Measurement: RiboLace/Ribo-Calibration for active ribosome quantification
- Stability Integration: Avoiding collision-induced decay through balanced loading
“The era of maximal ribosome density is over – therapeutic mRNA now demands optimized loading where translation efficiency dances in equilibrium with transcript longevity.”
— mRNA Therapeutics ReviewFuture advancements will focus on self-regulating mRNAs that dynamically adjust ribosome load based on cellular conditions.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.
- Stress-Responsive UTRs
